Posts tagged "python":
Python Inception
At most companies I have worked for, there was some internal Python code base that relied on an old version of Python. But especially for data science, I'd often want to execute that code from an up-to-date Jupyter Notebook, to do some analysis on results.
When this happened last time, I decided to do something about it. Here's a Jupyter cell magic that executes the cell's code in a different Python, pipes out all of STDOUT and STDERR, and imports any newly created variables into the host Python. Use it like this:
%%py_magic /old/version/of/python import this truth = 42
When this cell executes, you will see the Zen of Python in your output, just as if you had import this in the host Python, and the variable truth will now be 42 in the host Python.
To get this magic, execute the following code in a preceding cell:
import subprocess import sys import pickle import textwrap from IPython.core.magic import needs_local_scope, register_cell_magic @register_cell_magic @needs_local_scope def py_magic(line, cell, local_ns=None): proc = subprocess.Popen([line or 'python'], stdin=subprocess.PIPE, stdout=subprocess.PIPE, stderr=subprocess.PIPE, encoding='UTF8') # send a preamble to the client python, and remember all pre-existing local variable names: proc.stdin.write(textwrap.dedent(""" import pickle as _pickle import types as _types _names_before = [k for k, v in locals().items()] + ['_f', '_names_before'] try: """)) # send the cell's contents, indented to run in the try: for line in cell.splitlines(): proc.stdin.write(" " + line + "\n") # indent! # send a postamble that pickles all new variables or thrown exceptions: proc.stdin.write(textwrap.dedent(""" # save results to result.pickle except Exception as exc: with open('result.pickle', 'wb') as _f: _pickle.dump({'type':'error', 'value': exc}, _f) else: with open('result.pickle', 'wb') as _f: _values = {k:v for k, v in locals().items() if not isinstance(v, _types.ModuleType) and not k in _names_before} _safe_values = {} # skip any unpickleable variables for k, v in _values.items(): try: _pickle.dumps(v) except Exception as _exc: print(f'skipping dumping {k} because {_exc}') else: _safe_values[k] = v _pickle.dump({'type':'result', 'value': _safe_values}, _f) finally: quit() """)) # print any captured stdout or stderr: stdout, stderr = proc.communicate() if stdout: print(stdout, file=sys.stdout) if stderr: print(stderr, file=sys.stderr) # load new local variables or throw error: try: with open('result.pickle', 'rb') as f: result = pickle.load(f) if result['type'] == 'error': raise result['value'] elif result['type'] == 'result': for key, value in result['value'].items(): try: local_ns[key] = value except Exception as exc: print(f"skipping loading {key} because {exc}") finally: pathlib.Path('result.pickle').unlink() # remove temporary file del py_magic # otherwise the function overwrites the magic
I love how this sort of trickery is relatively easy in Python. Also, this is the first time I've used a try with except, else, and finally.
File Parsing with Python Video Tutorial
This video series was produced in the spring of 2020, during the COVID19-pandemic, when all lectures had to be held electronically, without physical attendance. It is a tutorial, in German, for parsing text files, and basic unit testing.
If the videos are too slow, feel free to speed them up by right-clicking, and adjusting play speed (Firefox only, as far as I know).
You may also download the videos and share them with your friends. Please do not upload them to social media or YouTube, but link to this website instead. If you want to modify them or create derivative works, please contact me.

The Qt for Python Video Tutorial by Bastian Bechtold is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
1 Intro
Prerequisites: A basic understanding of Python, and a working installation of python ≥3.6.
An overview over the topics discussed in the rest of the videos, and installation of pytest.
2 INI: First Steps
Basic setup, and our first test.
Code: inifile_1.py and inifile_test_1.py
3 INI: Sections
Parsing INI sections.
Code: inifile_2.py and inifile_test_2.py
4 INI: Variables
Parsing INI variable assignments.
Code: inifile_3.py and inifile_test_3.py
5 INI: Bugfixes and Integration Tests
Parsing difficult values, and comments.
Code: inifile_4.py and inifile_test_4.py
6 INI: Test the Tests
Tests can be wrong, too.
7 CSV: First Prototype
A simple parser for values without quotes.
Code: csvfile_1.py and csvfile_test_1.py
8 CSV: Quotes
Parsing quoted values makes everything harder.
Code: csvfile_2.py and csvfile_test_2.py
9 CSV: A Few More Features
Comments and a choice of separators.
Code: csvfile_3.py and csvfile_test_3.py
10 JSON: Keyword Parser
Parsing the simplest of JSON expressions.
Code: jsonfile_1.py and jsonfile_test_1.py
11 JSON: Strings
Parsing JSON strings is not as simple as it seems.
Code: jsonfile_2.py and jsonfile_test_2.py
12 JSON: Numbers
Numbers in JSON.
Code: jsonfile_3.py and jsonfile_test_3.py
13 JSON: Data Structures
The rest of JSON: Objects and Arrays.
Code: jsonfile_4.py and jsonfile_test_4.py
14 Regular Expressions 1
How to parse parts of INI files with regular expressions.
Code: inifile_regex.py
15 Regular Expressions 2
How to parse parts of JSON files with regular expressions.
Code: jsonfile_regex.py
16 Wrapup
A summary of the topics discussed.
Qt for Python Video Tutorial
This video series was produced in the spring of 2020, during the COVID19-pandemic, when all lectures had to be held electronically, without physical attendance. It is a tutorial, in German, for building a small Qt GUI that visualizes the ongoing pandemic on a world map.
If the videos are too slow, feel free to speed them up by right-clicking, and adjusting play speed (Firefox only, as far as I know).
You may also download the videos and share them with your friends. Please do not upload them to social media or YouTube, but link to this website instead. If you want to modify them or create derivative works, please contact me.

The Qt for Python Video Tutorial by Bastian Bechtold is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
Update: As of April 2022, all code examples have been updated to use PySide6. In particular, this changes the imports, replaces app.exec_() with app.exec(), and replaces mouseEvent.pos() with mouseEvent.position().toPoint() (see note in map11.py:).
1 Intro
Prerequisites: A basic understanding of Python, and a working installation of python ≥3.4.
An overview over the topics discussed in the rest of the videos, and installation of Qt for Python and Pandas.
2 Hello World
Our first GUI program, a window with a text label.
Code: map2.py
3 Main Window
Create a QMainWindow, and build some structure for later episodes.
Code: map3.py
4 Layouts
Position a QLabel and a QPushButton side by side, using layouts.
Code: map4.py
5 Signals and Slots
Make the button change the label's text if clicked.
Code: map5.py
6 Loading Data
7 Drawing the Map
Draw a world map into a QGraphicsScene.
Code: map7.py
9 Resize Event
Resize the map when the window size changes, by overloading resizeEvent.
Code: map9.py
10 Mouse Tracking
Highlight the country under the mouse.
Code: map10.py
11 Custom Signal
Respond to clicks of a country.
Code: map11.py
12 Addendum
Improve the code by cutting out a middle man.
Code: map12.py
13 Pandas
A quick introduction to Pandas.
Data: covid19.csv
14 Pandas Integration
Load the COVID19 dataset and print some stats.
Code: map14.py
15 Model View Tables
Display the COVID19 dataset in a QTableView.
Code: map15.py
16 Table Header Data
Fill in the table headers from the dataset.
Code: map16.py
17 Country Selection
Show only a subset of the dataset when a country is clicked.
Code: map17.py
18 Cleanup
Summary, and a few finishing touches.
Code: map18.py
Cool Python Libraries: TQDM and Resampy
In my recent post about appreciation for open source software, I mentioned that we should praise our open source heros more often. So here are two lesser-known libraries that I use daily, and which are unabashedly awesome:
TQDM
TQDM draws text progress bars for long-running processes, simply by wrapping your iterator in tqdm(iterator). And this, alone, would be awesome. But, TQDM is one of those libraries that aren't just a good idea, but then go the extra mile, and add fantastic documentation, contingencies for all kinds of weird use cases, and integration with notebooks and GUIs.
I use TQDM all the time, for running my scientific experiments and data analysis, and it just works. For long-running tasks, I recommend using tqdm(iterator, smoothing=0, desc='calculating'), which adds a meaningful description to the progress bar, and an accurate runtime estimate.
Resampy
Resampy resamples numpy signals. Resample your data with resample(signal, old_samplerate, new_samplerate). Just like with TQDM, this simple interface hides a lot of complexity and flexibility under the hood, yet remains conceptually simple and easy to use.
But beyond simplicity, resampy uses a clever implementation that is a far cry better than scipy.signal.resample, while still being easy to install and fast. For a more thorough comparison of resampling algorithms, visit Joachim Thiemann's blog.
PyEnv is the new Conda
How to install Python? If your platform has a package manager, you might be tempted to use that to install Python. But I don't like that. That version is often outdated, and you risk messing with an integral part of your operating system. Instead, I like to install a separate Python in my home directory. I used to use Anaconda (or WinPython, or EPD) to do this. But now there is a better way: PyEnv
The thing is: PyEnv installs (any version of) Python. That's all it does.
So why would I choose PyEnv over the more popular Anaconda? Because Anaconda is a Python distribution, a package manager, an environment manager, and a platform for paid packages. In the past, they once did break pip because they wanted to promote conda instead. Some features of conda require a login, some require a paid subscription. When you install packages through conda, you get binaries and source code from anaconda's servers, not the official packages from PyPi, which might or might not be up-to-date and feature-complete. For every package you install, you have to make a choice of using pip or conda, and the same goes for specifying your dependencies.
As an aside, many of these complaints are just as true for package-manager-provided Python packages (which often break pip, too!). Just like Anaconda, package managers want to be the true and only source of packages, and don't like to interact with Python's own package manager.
In contrast, with PyEnv, you install a Python. This can be a version of CPython, PyPy, IronPython, Jython, Pyston, stackless, miniconda, or even Anaconda. It downloads the sources from the official repos, and compiles them on your machine [1]. Plus, it provides an easy and transparent way of switching between installed versions (including any system-installed versions). After that, you use Python's own venv and pip.
I find this process much simpler, and easier to manage, because it relies on small, orthogonal tools (pyenv, venv, pip) instead of one integrated conda that kind of does everything. I also like to use these official tools and packages instead of conda's parallel universe of mostly-open, mostly-free, mostly-standard replacements.
Mind you, conda solved real problems back in the day (binary package distributions, Python version management, and environment management), and arguably still does (MKL et al, paid packages). But ever since wheels became ubiquitous and painless, and virtualenv was integrated into Python, and the development of PyEnv, these issues now have better solutions, and conda is no longer needed for my applications.
[1] the downside of compilation is: no Windows support.
Pip is great
Installing Python packages used to be a pain. Very surprisingly, this is no longer the case. Nowadays, pip install whatever will reliably install pretty much anything without any trouble.
To recap, the problem is that many Python packages rely on C code, which needs to be compiled before installation. In the past, this burden was mostly on the user. Depending on the user's knowledge of C and compilers, and the user's operating system, this could become almost arbitrarily hairy.
This problem was solved, to some extent, using pre-compiled packages, first as binary installers on the package websites, then pre-packaged Python distributions, and later through third-party package managers such as conda.
This worked well, but it fractured the ecosystem into several different mostly-compatible package sources. This was no big problem, but users had to decide on a package-by-package basis whether to download an installer, use conda, or use pip.
But I am happy to announce that these dark days are behind us now. Pip now automatically installs binary Python packages called wheels, and most package developers do now provide wheels on PyPI. This is so obviously superior to the past that many high-profile packages don't even provide binary installers any more.
For me, this means I don't have to rely on conda any longer. I don't have to keep a mental list of which packages to install though conda and which packages to install through pip any longer. I can go back to using virtualenv instead of conda-envs1 again. I don't have to tell students to download pre-packaged Python distributions any longer. Pip is great!
Footnotes:
Not because I dislike conda, just to simplify my development environment.
Transplant, revisited
A few months ago, I talked about the performance of calling Matlab from Python. Since then, I implemented a few optimizations that make working with Transplant a lot faster:
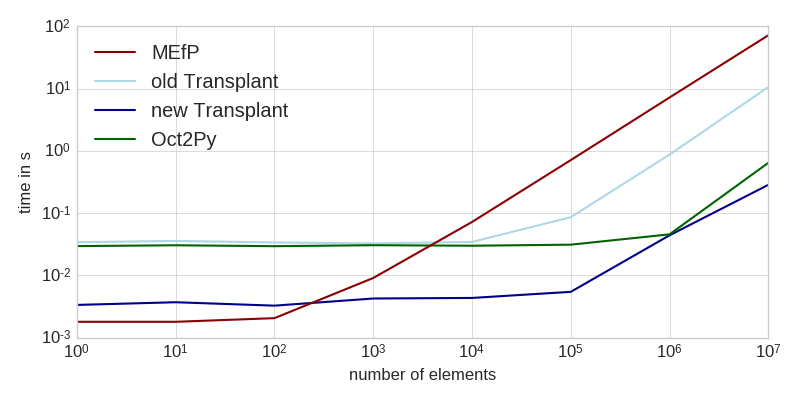
The workload consisted of generating a bunch of random numbers (not included in the times), and sending them to Matlab for computation. This task is entirely dominated by the time it takes to transfer the data to Matlab (see table at the end for intra-language benchmarks of the same task).
As you can see, the new Transplant is significantly faster for small workloads, and still a factor of two faster for larger amounts of data. It is now almost always a faster solution than the Matlab Engine for Python (MEfP) and Oct2Py. For very large datasets, Oct2Py might be preferable, though.
This improvement comes from three major changes: Matlab functions are now returned as callable objects instead of ad-hoc functions, Transplant now uses MsgPack instead of JSON, and loadlibrary instead of a Mex file to call into libzmq. All of these changes are entirely under the hood, though, and the public API remains unchanged.
The callable object thing is the big one for small workloads. The advantage is that the objects will only fetch documentation if __doc__ is actually asked for. As it turns out, running help('funcname') for every function call is kind of a big overhead.
Bigger workloads however are dominated by the time it takes Matlab to decode the data. String parsing is very slow in Matlab, which is a bad thing indeed if you're planning to read a couple hundred megabytes of JSON. Thus, I replaced JSON with MsgPack, which eliminates the parsing overhead almost entirely. JSON messaging is still available, though, if you pass msgformat='json' to the constructor. Edit: Additionally, binary data is no longer encoded as base64 strings, but passed directly through MsgPack. This yields about a ten-fold performance improvement, especially for larger data sets.
Lastly, I rewrote the ZeroMQ interaction to use loadlibrary instead of a Mex file. This has no impact on processing speed at all, but you don't have to worry about compiling that C code any more.
Oh, and Transplant now works on Windows!
Here is the above data again in tabular form:
| Task | New Transplant | Old Transplant | Oct2Py | MEfP | Matlab | Numpy | Octave |
|---|---|---|---|---|---|---|---|
| startup | 4.8 s | 5.8 s | 11 ms | 4.6 s | |||
| sum(randn(1,1)) | 3.36 ms | 34.2 ms | 29.6 ms | 1.8 ms | 9.6 μs | 1.8 μs | 6 μs |
| sum(randn(1,10)) | 3.71 ms | 35.8 ms | 30.5 ms | 1.8 ms | 1.8 μs | 1.8 μs | 9 μs |
| sum(randn(1,100)) | 3.27 ms | 33.9 ms | 29.5 ms | 2.06 ms | 2.2 μs | 1.8 μs | 9 μs |
| sum(randn(1,1000)) | 4.26 ms | 32.7 ms | 30.6 ms | 9.1 ms | 4.1 μs | 2.3 μs | 12 μs |
| sum(randn(1,1e4)) | 4.35 ms | 34.5 ms | 30 ms | 72.2 ms | 25 μs | 5.8 μs | 38 μs |
| sum(randn(1,1e5)) | 5.45 ms | 86.1 ms | 31.2 ms | 712 ms | 55 μs | 38.6 μs | 280 μs |
| sum(randn(1,1e6)) | 44.1 ms | 874 ms | 45.7 ms | 7.21 s | 430 μs | 355 μs | 2.2 ms |
| sum(randn(1,1e7)) | 285 ms | 10.6 s | 643 ms | 72 s | 3.5 ms | 5.04 ms | 22 ms |
Updating the Matplotlib Font Cache
When publishing papers or articles, I want my plots to integrate with the text surrounding them. I want them to use the correct font size, and the correct font.
This is easy to do with Matplotlib:
import matplotlib matplotlib.rcParams['font.size'] = 12 matplotlib.rcParams['font.family'] = 'Calibri'
However, sometimes, Matplotlib won't find the correct, even though it is clearly installed. This happens when Matplotlib's internal font cache is out of date.
To refresh the font cache, use
matplotlib.font_manager._rebuild()
Happy Plotting!
Calling Matlab from Python
For my latest experiments, I needed to run both Python functions and Matlab functions as part of the same program. As I noted earlier, Matlab includes the Matlab Engine for Python (MEfP), which can call Matlab functions from Python. Before I knew about this, I created Transplant, which does the very same thing. So, how do they compare?
Usage
As it's name suggests, Matlab is a matrix laboratory, and matrices are the most important data type in Matlab. Since matrices don't exist in plain Python, the MEfP implements it's own as matlab.double et al., and you have to convert any data you want to pass to Matlab into one of those. In contrast, Transplant recognizes the fact that Python does in fact know a really good matrix engine called Numpy, and just uses that instead.
Matlab Engine for Python | Transplant
---------------------------------------|---------------------------------------
import numpy | import numpy
import matlab | import transplant
import matlab.engine |
|
eng = matlab.engine.start_matlab() | eng = transplant.Matlab()
numpy_data = numpy.random.randn(100) | numpy_data = numpy.random.randn(100)
list_data = numpy_data.tolist() |
matlab_data = matlab.double(list_data) |
data_sum = eng.sum(matlab_data) | data_sum = eng.sum(numpy_data)
Aside from this difference, both libraries work almost identical. Even the handling of the number of output arguments is (accidentally) almost the same:
Matlab Engine for Python | Transplant
---------------------------------------|---------------------------------------
eng.max(matlab_data) | eng.max(numpy_data)
>>> 4.533 | >>> [4.533 537635]
eng.max(matlab_data, nargout=1) | eng.max(numpy_data, nargout=1)
>>> 4.533 | >>> 4.533
eng.max(matlab_data, nargout=2) | eng.max(numpy_data, nargout=2)
>>> (4.533, 537635.0) | >>> [4.533 537635]
Similarly, both libraries can interact with Matlab objects in Python, although the MEfP can't access object properties:
Matlab Engine for Python | Transplant
---------------------------------------|---------------------------------------
f = eng.figure() | f = eng.figure()
eng.get(f, 'Position') | eng.get(f, 'Position')
>>> matlab.double([[ ... ]]) | >>> array([[ ... ]])
f.Position | f.Position
>>> AttributeError | >>> array([[ ... ]])
There are a few small differences, though:
- Function documentation in the MEfP is only available as
eng.help('funcname'). Transplant will populate a function's__doc__, and thus documentation tools like IPython's?operator just work. - Transplant converts empty matrices to
None, whereas the MEfP represents them asmatlab.double([]). - Transplant represents
dictascontainers.Map, while the MEfP usesstruct(the former is more correct, the latter arguable more useful). - If the MEfP does not know
nargout, it assumesnargout=1. Transplant usesnargout(func)or returns whatever the function writes intoans. - The MEfP can't return non-scalar structs, such as the return value of
whos. Transplant can do this. - The MEfP can't return anonymous functions, such as
eng.eval('@(x, y) x>y'). Transplant can do this.
Performance
The time to start a Matlab instance is shorter in MEfP (3.8 s) than in Transplant (6.1 s). But since you're doing this relatively seldomly, the difference typically doesn't matter too much.
More interesting is the time it takes to call a Matlab function from Python. Have a look:

This is running sum(randn(n,1)) from Transplant, the MEfP, and in Matlab itself. As you can see, the MEfP is a constant factor of about 1000 slower than Matlab. Transplant is a constant factor of about 100 slower than Matlab, but always takes at least 0.05 s.
There is a gap of about a factor of 10 between Transplant and the MEfP. In practice, this gap is highly significant! In my particular use case, I have a function that takes about one second of computation time for an audio signal of ten seconds (half a million values). When I call this function with Transplant, it takes about 1.3 seconds. With MEfP, it takes 4.5 seconds.
Transplant spends its time serializing the arguments to JSON, sending that JSON over ZeroMQ to Matlab, and parsing the JSON there. Well, to be honest, only the parsing part takes any significant time, overall. While it might seem onerous to serialize everything to JSON, this architecture allows Transplant to run over a network connection.
It is a bit baffling to me that MEfP manages to be slower than that, despite being written in C. Looking at the number of function calls in the profiler, the MEfP calls 25 functions (!) on each value (!!) of the input data. This is a shockingly inefficient way of doing things.
TL;DR
It used to be very difficult to work in a mixed-language environment, particularly with one of those languages being Matlab. Nowadays, this has thankfully gotten much easier. Even Mathworks themselves have stepped up their game, and can interact with Python, C, Java, and FORTRAN. But their interface to Python does leave something to be desired, and there are better alternatives available.
If you want to try Transplant, just head over to Github and use it. If you find any bugs, feature requests, or improvements, please let me know in the Github issues.
Numpy Broadcasting Rules
They say that all arithmetic operations in Numpy behave like their element-wise cousins in Matlab. This is wrong, and seriously tripped me up last week.
In particular, this is what happens when you multiply an array with a matrix[1] in Numpy:
[[ 1], [[1, 2, 3], [[ 1, 2, 3],
[ 10], * [4, 5, 6], = [ 40, 50, 60],
[100]] [7, 8, 9]] [700, 800, 900]]
[ 1, 10, 100] [[1, 2, 3], [[ 1, 20, 300],
OR * [4, 5, 6], = [ 4, 50, 600],
[[ 1, 10, 100]] [7, 8, 9]] [ 7, 80, 900]]
They behave as if each row was evaluated separately, and singular dimensions are repeated where necessary. It helps to think about them as row-wise, instead of element-wise. This is particularly important in the second example, where the whole 1d-array is multiplied with every row of the 2d-array.
Note that this is not equivalent to multiplying every element as in [a[n]*b[n] for n in range(len(a))]. I guess that's why this is called broadcasting, and not element-wise.
[1] "matrix" here refers to a 2-d numpy.array. There is also a numpy.matrix, where multiplication is matrix multiplication, but this is not what I'm talking about.
Python Numeric Performance
Recently, I was working on a dynamic programming algorithm that involves a lot of number crunching in nested loops. The algorithm looks like this:
def y_change_probability_python(oct_per_sec):
""" ... """
b = 1.781
mu = -0.301
return 1/(2*b)*math.exp(-abs(oct_per_sec-mu)/b)
def y_idx_range_python(y_idx, max_y_factor, height):
""" ... """
y = (y_idx/height)*(max_y-min_y)+min_y
y_lo = max(y/max_y_factor, min_y)
y_hi = min(y*max_y_factor, max_y)
y_lo_idx = int((y_lo-min_y)/(max_y-min_y)*height)
y_hi_idx = int((y_hi-min_y)/(max_y-min_y)*height)
return y_lo_idx, y_hi_idx
def find_tracks_python(cost_matrix, delta_x):
""" ... """
tracks = np.zeros(correlogram.shape, dtype=np.int64)
cum_cost = np.zeros(correlogram.shape)
max_y_factor = (2**5)**(delta_x)
height = correlogram.shape[1]
probabilities = np.empty((height, height))
for y_idx in range(height):
y = (y_idx/height)*(max_y-min_y)+min_y
for y_pre_idx in range(*y_idx_range_numba(y_idx, max_y_factor, height)):
y_pre = (y_pre_idx/height)*(max_y-min_y)+min_y
doubles_per_x = math.log2((y/y_pre)**(1/delta_x))
probabilities[y_idx, y_pre_idx] = y_change_probability_numba(doubles_per_x)
for x_idx, cost_column in enumerate(cost_matrix):
if x_idx == 0:
cum_cost[x_idx] = cost_column
continue
for y_idx, cost in enumerate(cost_column):
for y_pre_idx in range(*y_idx_range_numba(y_idx, max_y_factor, height)):
weighted_cum_cost = cum_cost[x_idx-1, y_pre_idx] + cost*probabilities[y_idx, y_pre_idx]
if weighted_cum_cost > cum_cost[x_idx, y_idx]:
cum_cost[x_idx, y_idx] = weighted_cum_cost
tracks[x_idx, y_idx] = y_pre_idx
cum_cost[x_idx, y_idx] = cum_cost[x_idx-1, tracks[x_idx, y_idx]] + cost
return tracks, cum_cost
I'm not going into the details of what this algorithm does, but note that it iterates over every column and row of the matrix cost_matrix, and then iterates over another range previous_y_range for each of the values in cost_matrix. On the way, it does a lot of basic arithmetic and some algebra.
The problem is, this is very slow. For a \(90 \times 200\) cost_matrix, this takes about 260 ms. Lots of loops? Lots of simple mathematics? Slow? That sounds like a perfect match for Numpy!
If you can express your code in terms of linear algebra, Numpy will execute them in highly-optimized C code. The problem is, translating loops into linear algebra is not always easy. In this case, it took some effort:
def y_change_probability_numpy(doubles_per_x):
""" ... """
b = 1.781
mu = -0.301
return 1/(2*b)*np.exp(-np.abs(doubles_per_x-mu)/b)
def find_frequency_tracks_numpy(cost_matrix, delta_x):
""" ... """
tracks = np.zeros(cost_matrix.shape, dtype=np.int)
cum_cost = np.zeros(cost_matrix.shape)
max_y_factor = (2**5)**(delta_t) # allow at most 5 octaves per second (3 sigma)
height = cost_matrix.shape[1]
# pre-allocate probabilities matrix as minus infinity. This matrix
# will be sparsely filled with positive probability values, and
# empty values will have minus infinite probability.
probabilities = -np.ones((height, height))*np.inf
for y_idx in range(probabilities.shape[0]):
y = (y_idx/height)*(max_y-min_y)+min_y
y_pre_idx = np.arange(int((max(y/max_y_factor, min_y)-min_y)/(max_y-min_y)*height),
int((min(y*max_y_factor, max_y)-min_y)/(max_y-min_y)*height))
y_pre = (y_pre_idx/height)*(max_y-min_y)+min_y
doubles_per_x = np.log2((y/y_pre)**(1/delta_x))
probabilities[y_idx, y_pre_idx] = y_change_probability(doubles_per_x)
cum_cost[0] = cost_matrix[0]
for x_idx in range(1, len(cost_matrix)):
cost_column = cost_matrix[x_idx:x_idx+1] # extract cost_column as 2d-vector!
weighted_cum_cost = cum_cost[x_idx-1] + cost_column.T*probabilities
tracks[x_idx] = np.argmax(weighted_cum_cost, axis=1)
cum_cost[x_idx] = cum_cost[x_idx-1, tracks[x_idx]] + cost_column
return tracks, cum_corrs
This code does not look much like the original, but calculates exactly the same thing. This takes about 15 ms for a \(90 \times 200\) cost_matrix, which is about 17 times faster than the original code! Yay Numpy! And furthermore, this code is arguably more readable than the original, since it is written at a higher level of abstraction.
Another avenue for performance optimization is Numba. Numba applies dark and powerful magic to compile humble Python functions into blazingly fast machine code. It is proper magic, if you ask me. Simply add an innocuous little decorator to your functions, and let Numba do it's thing. If all goes well, your code will work just as before, except with unheard-of performance:
@jit(numba.float64(numba.float64), nopython=True)
def y_change_probability_numba(doubles_per_x):
...
@jit((numba.int64, numba.float64, numba.int64), nopython=True)
def y_idx_range_numba(y_idx, max_y_factor, height):
...
@jit((numba.float64[:,:], numba.float64), nopython=True)
def find_tracks_numba(cost_matrix, delta_t):
...
However, Numba is no silver bullet, and does not support all of Python yet. In the present case, it is missing support for enumerate for Numpy matrices. Thus, I had to rewrite the first two loops like this:
# python version
for x_idx, cost_column in enumerate(cost_matrix):
...
# numba version
for x_idx in range(len(cost_matrix)):
cost_column = cost_matrix[x_idx]
...
Another area that proved problematic is N-D slice writing. Instead of using expressions like m1[x,y:y+3] = m2, you have to write for idx in range(3): m1[x,y+idx] = m2[idx]. Not a difficult transformation, but it basically forced me to unroll all the nice vectorized code of the Numpy version back to their original pure-Python form. That said, Numba is getting better and better, and many constructs that used to be uncompilable (yield) are not a problem any more.
Anyway, with that done, the above code went down from 260 ms to 2.2 ms. This is a 120-fold increase in performance, and still seven times faster than Numpy, with minimal code changes. This is proper magic!
So why wouldn't you just always use Numba? After all, when it comes down to raw performance, Numba is the clear winner. The big difference between performance optimization using Numpy and Numba is that properly vectorizing your code for Numpy often reveals simplifications and abstractions that make it easier to reason about your code. Numpy forces you to think in terms of vectors, matrices, and linear algebra, and this often makes your code more beautiful. Numba on the other hand often requires you to make your code less beautiful to conform to it's subset of compilable Python.
Transplant
In academia, a lot of programming is done in Matlab. Many very interesting algorithms are only available in Matlab. Personally, I prefer to use tools that are more widely applicable, and less proprietary, than Matlab. My weapon of choice at the moment is Python.
But I still need to use Matlab code. There are a few ways of interacting with Matlab out there already. Most of them focus on being able to eval strings in Matlab. Boring. The most interesting one is mlab, a full-fledget bridge between Python and Matlab! Had I found this earlier, I would probably not have written my own.
But write my own I did: Transplant. Transplant is a very simple bridge for calling Matlab functions from Python. Here is how you start Matlab from Python:
import transplant matlab = transplant.Matlab()
This matlab object starts a Matlab interpreter in the background and connects to it. You can call Matlab functions on it!
matlab.eye(3)
>>> array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., 1.]])
As you can see, Matlab matrices are converted to Numpy matrices. In contrast to most other Python/Matlab bridges, matrix types are preserved1:
matlab.randi(255, 1, 4, 'uint8') >>> array([[246, 2, 198, 209]], dtype=uint8)
All matrix data is actually transferred in binary, so both Matlab and Python work on bit-identical data. This is very important if you are working with precise data! Most other bridges do some amount of type conversion at this point.
This alone accounts for a large percentage of Matlab code out there. But not every Matlab function can be called this easily from Python: Matlab functions behave differently depending the number of output arguments! To emulate this in Python, every function has a keyword argument nargout 2. For example, the Matlab function max by default returns both the maximum value and the index of that value. If given nargout=1 it will only return the maximum value:
data = matlab.randn(1, 4) matlab.max(data) >>> [1.5326, 3] # Matlab: x, n = max(...) matlab.max(data, nargout=1) >>> 1.5326 # Matlab: x = max(...)
If no nargout is given, functions behave according to nargout(@function). If even that fails, they return the content of ans after their execution.
Calling Matlab functions is the most important feature of Transplant. But there is a more:
You can save/retrieve variables in the global workspace:
matlab.value = 5 # Matlab: value = 5 x = matlab.value # Matlab: x = value
You van eval some code:
matlab.eval('class(value)') >>> ans = >>> >>> double >>>- The help text for functions is automatically assigned as docstring. In IPython, this means that
matlab.magic?displays the same thinghelp magicwould display in Matlab.
Under the hood, Transplant is using a very simple messaging protocol based on 0MQ, JSON, and some base64-encoded binary data. Sadly, Matlab can deal with none of these technologies by itself. Transplant therefore contains a full-featured JSON parser/serializer and base64 encoder/decoder in pure Matlab. It also contains a minimal mex-file for interfacing with 0MQ.
There are a few JSON parsers available for Matlab, but virtually all of them try parse JSON arrays as matrices. This means that these parsers have no way of differentiating between a list of vectors and a matrix (want to call a function with three vectors or a matrix? No can do). Transplant's JSON parser parses JSON arrays as cell arrays and JSON objects as structs. While somewhat less convenient in general, this is a much better fit for transferring data structures between programming languages.
Similarly, there are a few base64 encoders available. Most of them actually use Matlab's built-in Java interface to encode/decode base64 strings. I tried this, but it has two downsides: Firstly, it is pretty slow for short strings since the data has to be copied over to the Java side and then back. Secondly, it is limited by the Java heap space. I was not able to reliably encode/decode more than about 64 Mb using this3. My base64 encoder/decoder is written in pure Matlab, and works for arbitrarily large data.
All of this has been about Matlab, but my actual goal is bigger: I want transplant to become a library for interacting between more than just Python and Matlab. In particular, Julia and PyPy would be very interesting targets. Also, it would be useful to reverse roles and call Python from Matlab as well! But that will be in the future.
For now, head over to Github.com/bastibe/transplant and have fun! Also, if you find any bugs or have any suggestions, please open an issue on Github!
Footnotes:
Sound in Python
Have you ever wanted to work with audio data in Python? I know I do. I want to record from the microphone, I want to play sounds. I want to read and write audio files. If you ever tried this in Python, you know it is kind of a pain.
It's not for a lack of libraries though. You can read sound files using wave, SciPy provides scipy.io.wavfile, and there is a SciKit called scikits.audiolab. And except for scikits.audiolab, these return the data as raw bytes. Like, they parse the WAVE header and that is great and all, but you still have to decode your audio data yourself.
The same thing goes for playing/recording audio: PyAudio provides nifty bindings to portaudio, but you still have to decode your raw bytes by hand.
But really, what I want is something different: When I record from the microphone, I want to get a NumPy array, not bytes. You know, something I can work with! And then I want to throw that array into a sound file, or play it on a different sound card, or do some calculations on it!
So one fateful day, I was sufficiently frustrated with the state of things that I set out to create just that. Really, I only wanted to play around with cffi, but that is beside the point.
So, lets read some audio data, shall we?
import soundfile
data = soundfile.read('sad_song.wav')
done. All the audio data is now available as a NumPy array in data. Just like that.
Awesome, isn't it?
OK, that was easy. So let's read only the first and last 100 frames!
import soundfile
first = soundfile.read('long_song.flac', stop=100)
last = soundfile.read(start=-100)
This really only read the first and last bit. Not everything in between!
Note that at no point I did explicitly open or close a file! This is Python! We can do that! When the SoundFile object is created, it opens the file. When it goes out of scope, it closes the file. It's as simple as that. Or just use SoundFile in a context manager. That works as well.
Oh, but I want to use the sound card as well! I want to record audio to a file!
from pysoundcard import Stream
from pysoundfile import SoundFile, ogg_file, write_mode
with Stream() as s: # opens your default audio device
# This is supposed to be a new file, so specify it completely
f = SoundFile('happy_song.ogg', sample_rate=s.sample_rate,
channels=s.channels, format=ogg_file,
mode=write_mode)
f.write(s.read(s.sample_rate)) # one second
Read from the stream, write to a file. It works the other way round, too!
And that's really all there is to it. Working with audio data in Python is easy now!
Of course, there is much more you could do. You could create a callback function and be called every four[1] frames with new audio data to process. You could request your audio data as int16, because that would be totally awesome! You could use many different sound cards at the same time, and route stuff to and fro to your hearts desire! And you can run all this on Linux using ALSA or Jack, or on Windows using DirectSound or ASIO, or on Mac using CoreAudio[2]. And you already saw that you can read Wave files, OGG, FLAC or MAT-files[3].
You can download these libraries from PyPi, or use the binary Windows installers on Github. Or you can look at the source on Github (PySoundFile, PySoundCard), because Open Source is awesome like that! Also, you might find some bugs, because I haven't found them all yet. Then, I would like you to open an issue on Github. Or if have a great idea of how to improve things, please let me know as well.
UPDATE: It used to be that you could use indexing on SoundFile objects. For various political reasons, this is no longer the case. I updated the examples above accordingly.
[1] You can use any block size you want. Less than 4 frames per block can be really taxing for your CPU though, so be careful or you start dropping frames. [2] More precisely: Everything that portaudio supports. [3] More precisely: Everything that libsndfile supports.
Speeding up Matplotlib
For the record, Matplotlib is awesome! Its output looks amazing, it is extremely configurable and very easy to use. What more could you want?
Well… speed. If there is one thing I could criticize about Matplotlib, it is its relative slowness. To measure that, lets make a very simple line plot and draw some random numbers as quickly as possible:
import matplotlib.pyplot as plt
import numpy as np
import time
fig, ax = plt.subplots()
tstart = time.time()
num_plots = 0
while time.time()-tstart < 1:
ax.clear()
ax.plot(np.random.randn(100))
plt.pause(0.001)
num_plots += 1
print(num_plots)
On my machine, I get about 11 plots per second. I am using pause() here to update the plot without blocking. The correct way to do this is to use draw() instead, but due to a bug in the Qt4Agg backend, you can't use it there. If you are not using the Qt4Agg backend, draw() is supposedly the correct choice.
For a single plot, ten plots per second is not terrible. But then, this is really the simplest case possible, so ten frames per second in the simplest case probably means bad things for not so simple cases.
One thing that really takes time here is creating all the axes and text labels over and over again. So let's not do that.
Instead of calling clear() and then plot(), thus effectively deleting everything about the plot, then re-creating it for every frame, we can keep an existing plot and only modify its data:
fig, ax = plt.subplots()
line, = ax.plot(np.random.randn(100))
tstart = time.time()
num_plots = 0
while time.time()-tstart < 1:
line.set_ydata(np.random.randn(100))
plt.pause(0.001)
num_plots += 1
print(num_plots)
which yields about 26 plots per second. Not bad for a simple change like this. The downside is that the axes are not re-scaled any longer when the data changes. Thus, they won't change their limits based on the data any more.
Profiling this yields some interesting results:
| ncalls | tottime | percall | cumtime | percall | filename:lineno(function) |
| 15 | 0.167 | 0.011 | 0.167 | 0.011 | {built-in method sleep) |
The one function that uses the biggest chunk of runtime is sleep(), of all things. Clearly, this is not what we want. Delving deeper into the profiler shows that this is indeed happening in the call do pause(). Then again, I was wondering if using pause really was a great idea for performance…
As it turns out, pause() internally calls fig.canvas.draw(), then plt.show(), then fig.canvas.start_event_loop(). The default implementation of fig.canvas.start_event_loop() then calls fig.canvas.flush_events(), then sleeps for the requested time. To add insult to injury, it even insists on sleeping at least one hundredth of a second, which actually explains the profiler output of 0.167 seconds of sleep() for 15 calls very well.
Putting this all together now yields:
fig, ax = plt.subplots()
line, = ax.plot(np.random.randn(100))
tstart = time.time()
num_plots = 0
while time.time()-tstart < 1:
line.set_ydata(np.random.randn(100))
fig.canvas.draw()
fig.canvas.flush_events()
num_plots += 1
print(num_plots)
which now plots about 40 frames per second. Note that the call to show() mentioned earlier can be omitted since the figure is already on screen. flush_events() just runs the Qt event loop, so there is probably nothing to optimize there.
The only thing left to optimize now is thus fig.canvas.draw(). What this really is doing is drawing all the artists contained in the ax. Those artists can be accessed using ax.get_children(). For a simple plot like this, the artists are:
- the background
ax.patch - the line, as returned from the
plot()function - the spines
ax.spines - the axes
ax.xaxisandax.yaxis
What we can do here is to selectively draw only the parts that are actually changing. That is, at least the background and the line. To only redraw these, the code now looks like this:
fig, ax = plt.subplots()
line, = ax.plot(np.random.randn(100))
plt.show(block=False)
tstart = time.time()
num_plots = 0
while time.time()-tstart < 5:
line.set_ydata(np.random.randn(100))
ax.draw_artist(ax.patch)
ax.draw_artist(line)
fig.canvas.update()
fig.canvas.flush_events()
num_plots += 1
print(num_plots/5)
Note that you have to add fig.canvas.update() to copy the newly rendered lines to the drawing backend.
This now plots about 500 frames per second. Five hundred times per second! Frankly, this is quite amazing!
Note that since we are only redrawing the background and the line, some detail in the axes will be overwritten. To also draw the spines, use for spine in ax.spines.values(): ax.draw_artist(spine). To draw the axes, use ax.draw_artist(ax.xaxis) and ax.draw_artist(ax.yaxis). If you draw all of them, you get roughly the same performance as fig.canvas.draw(). The axes in particular are quite expensive.
There is also a way of drawing the complete figure once and copying the complete but empty background, then reinstating that and only plotting a new line on top of it. This is equally fast as the code above without any visual artifacts, but breaks if you resize the plot.
In conclusion, I am quite impressed with the flexibility of Matplotlib. Matplotlib by default values quality over performance. But if you really need the performance at some point, it is flexible and hackable enough to let you tweak it to your hearts content. Really, an amazing piece of technology!
EDIT: As it turns out, fig.canvas.blit(ax.bbox) is a bad idea since it leaks memory like crazy. What you should use instead is fig.canvas.update(), which is equally fast but does not leak memory.
A Python Primer for Matlab Users
Why would you want to use Python over Matlab?
- Because Python is free and Matlab is not.
- Because Python is a general purpose programming language and Matlab is not.
Let me qualify that a bit. Matlab is a very useful programming environment for numerical problems. For a very particular set of problems, Matlab is an awesome tool. For many other problems however, it is just about unusable. For example, you would not write a complex GUI program in Matlab, you would not write your blogging engine in Matlab and you would not write a web service in Matlab. You can do all that and more in Python.
Python as a Matlab replacement
The biggest strength of Matlab is its matrix engine. Most of the data you work with in Matlab are matrices and there is a host of functions available to manipulate and visualize those matrices. Python, by itself, does not have a convenient matrix engine. However, there are three packages (think Matlab Toolboxes) out there that will add this capability to Python:
- Numpy (the matrix engine)
- Scipy (matrix manipulation)
- Matplotlib (plotting)
You can either grab the individual installers for Python, Numpy, Scipy and Matplotlib from their respective websites, or get them pre-packaged from pythonxy() or EPD.
A 30,000 foot overview
Like Matlab, Python is interpreted, that is, there is no need for a compiler and code can be executed at any time as long as Python is installed on the machine. Also, code can be copied from one machine to another and will run without change.
Like Matlab, Python is dynamically typed, that is, every variable can hold data of any type, as in:
# Python
a = 5 # a number
a = [1, 2, 3] # a list
a = 'text' # a string
Contrast this with C, where you can not assign different data types to the same variable:
// C
int a = 5;
float b[3] = {1.0, 2.0, 3.0};
char c[] = "text";
Unlike Matlab, Python is strongly typed, that is, you can not add a number to a string. In Matlab, adding a single number to a string will convert that string into an array of numbers, then add the single number to each of the numbers in the array. Python will simply throw an error.
% Matlab
a = 'text'
b = a + 5 % [121 106 125 121]
# Python
a = 'text'
b = a + 5 # TypeError: Can't convert 'int' object to str implicitly
Unlike Matlab, every Python file can contain as many functions as you like. Basically, you can organize your code in as many files as you want. To access functions from other files, use import filename.
Unlike Matlab, Python is very quick to start. In fact, most operating systems automatically start a new Python process whenever you run a Python program and quit that process once the program has finished. Thus, every Python program behaves as if it indeed were an independent program. There is no need to wait for that big Matlab mother ship to start before writing or executing code.
Unlike Matlab, the source code of Python is readily available. Every detail of Python's inner workings is available to everyone. It is thus feasible and encouraged to actively participate in the development of Python itself or some add-on package. Furthermore, there is no dependence on some company deciding where to go next with Python.
Reading Python
When you start up Python, it is a rather empty environment. In order to do anything useful, you first have to import some functionality into your workspace. Thus, you will see a few lines of import statements at the top of every Python file. Moreover, Python has namespaces, so if you import numpy, you will have to prefix every feature of Numpy with its name, like this:
import numpy
a = numpy.zeros(10, 1)
This is clearly cumbersome if you are planning to use Numpy all the time. So instead, you can import all of Numpy into the global environment like this:
from numpy import *
a = ones(30, 1)
Better yet, there is a pre-packaged namespace that contains the whole Numpy-Scipy-Matplotlib stack in one piece:
from pylab import *
a = randn(100, 1)
plot(a)
show()
Note that Python does not plot immediately when you type plot(). Instead, it will collect all plotting information and only show it on the screen once you type show().
So far, the code you have seen should look pretty familiar. A few differences:
- No semicolons at the end of lines;
In order to print stuff to the console, use the
print()function instead. - No
endanywhere. In Python, blocks of code are identified by indentation and they always start with a colon like so:
sum = 0
for n in [1, 2, 3, 4, 5]:
sum = sum + n
print(sum)
- Function definitions are different.
They use the
defkeyword instead offunction. You don't have to name the output variable names in the definition and instead usereturn().
# Python
def abs(number):
if number > 0:
return number
else:
return -number
% Matlab
function [out] = abs(number)
if number > 0
out = number
else
out = -number
end
end
- There is no easy way to write out a list or matrix.
Since Python only gains a matrix engine by importing Numpy, it does not have a convenient way of writing arrays or matrices. This sounds more inconvenient than it actually is, since you are probably using mostly functions like
zeros()orrandn()anyway and those work just fine. Also, many places accept Python lists (like this[1, 2, 3]) instead of Numpy arrays, so this rarely is a problem. Note that you must use commas to separate items and can not use semicolons to separate lines.
# create a numpy matrix:
m = array([[1, 2, 3],
[4, 5, 6],
[7, 8, 9]])
# create a Python list:
l = [1 2 3]
- Arrays access uses brackets and is numbered from 0.
Thus, ranges exclude the last number (see below).
Mostly, this just means that array access does not need any
+1or-1when indexing arrays anymore.
a = linspace(1, 10, 10)
one = a[0]
two = a[1]
# "6:8" is a range of two elements:
a[6:8] = [70, 80] # <-- a Python list!
Common traps
- Array slicing does not copy.
a = array([1 2 3 4 5])
b = a[1:4] # [2 3 4]
b[1] = rand() # this will change a and b!
# make a copy like this:
c = array(a[1:4], copy=True) # copy=True can be omitted
c[1] = rand() # changes only c
- Arrays retain their data type.
You can slice them, you can dice them, you can do math on them, but a 16 bit integer array will never lose its data type. Use
new = array(old, dtype=double)to convert an array of any data type to the defaultdoubletype (like in Matlab).
# pretend this came from a wave file:
a = array([1000, 2000, 3000, 4000, 5000], dtype=int16)
a = a * 10 # int16 only goes to 32768!
# a is now [10000, 20000, 30000, -25536, -15536]
Going further
Now you should be able to read Python code reasonably well. Numpy, Scipy and Matplotlib are actually modeled after Matlab in many ways, so many functions will have a very similar name and functionality. A lot of the numerical code you write in Python will look very similar to the equivalent code in Matlab. For a more in-depth comparison of Matlab and Python syntax, head over to the Numpy documentation for Matlab users.
However, since Python is a general purpose programming language, it offers some more tools. To begin with, there are a few more data types like associative arrays, tuples (unchangeable lists), proper strings and a full-featured object system. Then, there is a plethora of add-on packages, most of which actually come with your standard installation of Python. For example, there are internet protocols, GUI programming frameworks, real-time audio interfaces, web frameworks and game development libraries. Even this very blog is created using a Python static site generator.
Lastly, Python has a great online documentation site including a tutorial, there are many books on Python and there is a helpful Wiki on Python. There is also a tutorial and documentation for Numpy, Scipy and Matplotlib.
A great way to get to know any programming language is to solve the first few problems on project euler.
Real Time Signal Processing in Python
Wouldn't it be nice if you could do real time audio processing in a convenient programming language? Matlab comes to mind as a convenient language for signal processing. But while Matlab is pretty fast, it is really only fast for algorithms that can be vectorized. In audio however, we have many algorithms that need knowledge about the previous sample to calculate the next one, so they can't be vectorized.
But this is not going to be about Matlab. This is going to be about Python. Combine Python with Numpy (and Scipy and Matplotlib) and you have a signal processing system very comparable to Matlab. Additionally, you can do real-time audio input/output using PyAudio. PyAudio is a wrapper around PortAudio and provides cross platform audio recording/playback in a nice, pythonic way. (Real time capabilities were added in 0.2.6 with the help of yours truly).
However, this does not solve the problem with vectorization. Just like Matlab, Python/Numpy is only fast for vectorizable algorithms. So as an example, let's define an iterative algorithm that is not vectorizable:
A Simple Limiter
A limiter is an audio effect that controls the system gain so that it does not exceed a certain threshold level. One could do this by simply cutting off any signal peaks above that level, but that sounds awful. So instead, the whole system gain is reduced smoothly if the signal gets too loud and is amplified back to its original gain again when it does not exceed the threshold any more. The important part is that the gain change is done smoothly, since otherwise it would introduce a lot of distortion.
If a signal peak is detected, the limiter will thus need a certain amount of time to reduce the gain accordingly. If you still want to prevent all peaks, the limiter will have to know of the peaks in advance, which is of course impossible in a real time system. Instead, the signal is delayed by a short time to give the limiter time to adjust the system gain before the peak is actually played. To keep this delay as short as possible, this "attack" phase where the gain is decreased should be very short, too. "Releasing" the gain back up to its original value can be done more slowly, thus introducing less distortion.
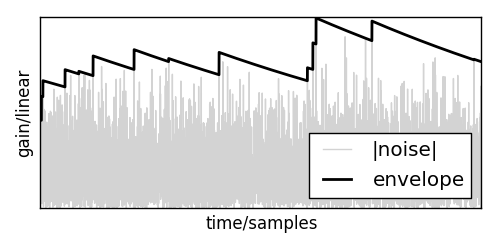
With that out of the way, let me present you a simple implementation of such a limiter. First, lets define a signal envelope \(e[n]\) that catches all the peaks and smoothly decays after them:
\[ e[n] = \max( |s[n]|, e[n-1] \cdot f_r ) \]
where \(s[n]\) is the current signal and \(0 < f_r < 1\) is a release factor.
If this is applied to a signal, it will create an envelope like this:
Based on that envelope, and assuming that the signal ranges from -1 to 1, the target gain \(g_t[n]\) can be calculated using
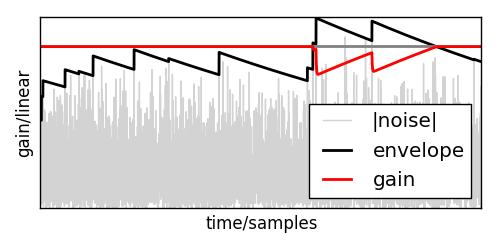
\[ g_t[n] = \begin{cases} 1 & e[n] < t \\ 1 + t - e[n] & e[n] > t \end{cases} \]
Now, the output gain \(g[n]\) can smoothly move towards that target gain using
\[ g[n] = g[n-1] \cdot f_a + g_t[n] \cdot (1-f_a) \]
where \(0 < f_a \ll f_r\) is the attack factor.
Here you can see how that would look in practice:
Zooming in on one of the limited section reveals that the gain is actually moving smoothly.
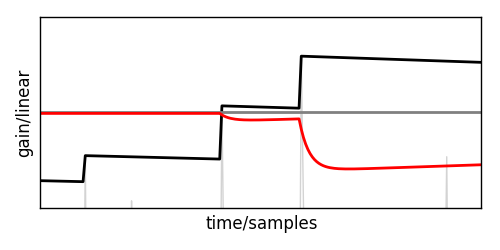
This gain can now be multiplied on the delayed input signal and will safely keep that below the threshold.
In Python, this might look like this:
class Limiter:
def __init__(self, attack_coeff, release_coeff, delay, dtype=float32):
self.delay_index = 0
self.envelope = 0
self.gain = 1
self.delay = delay
self.delay_line = zeros(delay, dtype=dtype)
self.release_coeff = release_coeff
self.attack_coeff = attack_coeff
def limit(self, signal, threshold):
for i in arange(len(signal)):
self.delay_line[self.delay_index] = signal[i]
self.delay_index = (self.delay_index + 1) % self.delay
# calculate an envelope of the signal
self.envelope *= self.release_coeff
self.envelope = max(abs(signal[i]), self.envelope)
# have self.gain go towards a desired limiter gain
if self.envelope > threshold:
target_gain = (1+threshold-self.envelope)
else:
target_gain = 1.0
self.gain = ( self.gain*self.attack_coeff +
target_gain*(1-self.attack_coeff) )
# limit the delayed signal
signal[i] = self.delay_line[self.delay_index] * self.gain
Note that this limiter does not actually clip all peaks completely, since the envelope for a single peak will have decayed a bit before the target gain will have reached it. Thus, the output gain will actually be slightly higher than what would be necessary to limit the output to the threshold. Since the attack factor is supposed to be significantly smaller than the release factor, this does not matter much though.
Also, it would probably be more useful to define the factors \(f_a\) and \(f_r\) in terms of the time they take to reach their target and the threshold \(t\) in dB FS.
Implementing audio processing in Python
A real-time audio processing framework using PyAudio would look like this:
(callback is a function that will be defined shortly)
from pyaudio import PyAudio, paFloat32
pa = PyAudio()
stream = pa.open(format = paFloat32,
channels = 1,
rate = 44100,
output = True,
frames_per_buffer = 1024,
stream_callback = callback)
while stream.is_active():
sleep(0.1)
stream.close()
pa.terminate()
This will open a stream, which is a PyAudio construct that manages input and output to/from one sound device. In this case, it is configured to use float values, only open one channel, play audio at a sample rate of 44100 Hz, have that one channel be output only and call the function callback every 1024 samples.
Since the callback will be executed on a different thread, control flow will continue immediately after pa.open(). In order to analyze the resulting signal, the while stream.is_active() loop waits until the signal has been processed completely.
Every time the callback is called, it will have to return 1024 samples of audio data. Using the class Limiter above, a sample counter counter and an audio signal signal, this can be implemented like this:
limiter = Limiter(attack_coeff, release_coeff, delay, dtype)
def callback(in_data, frame_count, time_info, flag):
if flag:
print("Playback Error: %i" % flag)
played_frames = counter
counter += frame_count
limiter.limit(signal[played_frames:counter], threshold)
return signal[played_frames:counter], paContinue
The paContinue at the end is a flag signifying that the audio processing is not done yet and the callback wants to be called again. Returning paComplete or an insufficient number of samples instead would stop audio processing after the current block and thus invalidate stream.is_active() and resume control flow in the snippet above.
Now this will run the limiter and play back the result. Sadly however, Python is just a bit too slow to make this work reliably. Even with a long block size of 1024 samples, this will result in occasional hickups and discontinuities. (Which the callback will display in the print(...) statement).
Speeding up execution using Cython
The limiter defined above could be rewritten in C like this:
// this corresponds to the Python Limiter class.
typedef struct limiter_state_t {
int delay_index;
int delay_length;
float envelope;
float current_gain;
float attack_coeff;
float release_coeff;
} limiter_state;
#define MAX(x,y) ((x)>(y)?(x):(y))
// this corresponds to the Python __init__ function.
limiter_state init_limiter(float attack_coeff, float release_coeff, int delay_len) {
limiter_state state;
state.attack_coeff = attack_coeff;
state.release_coeff = release_coeff;
state.delay_index = 0;
state.envelope = 0;
state.current_gain = 1;
state.delay_length = delay_len;
return state;
}
void limit(float *signal, int block_length, float threshold,
float *delay_line, limiter_state *state) {
for(int i=0; i<block_length; i++) {
delay_line[state->delay_index] = signal[i];
state->delay_index = (state->delay_index + 1) % state->delay_length;
// calculate an envelope of the signal
state->envelope *= state->release_coeff;
state->envelope = MAX(fabs(signal[i]), state->envelope);
// have current_gain go towards a desired limiter target_gain
float target_gain;
if (state->envelope > threshold)
target_gain = (1+threshold-state->envelope);
else
target_gain = 1.0;
state->current_gain = state->current_gain*state->attack_coeff +
target_gain*(1-state->attack_coeff);
// limit the delayed signal
signal[i] = delay_line[state->delay_index] * state->current_gain;
}
}
In contrast to the Python version, the delay line will be passed to the limit function. This is advantageous because now all audio buffers can be managed by Python instead of manually allocating and deallocating them in C.
Now in order to plug this code into Python I will use Cython. First of all, a "Cython header" file has to be created that declares all exported types and functions to Cython:
cdef extern from "limiter.h":
ctypedef struct limiter_state:
int delay_index
int delay_length
float envelope
float current_gain
float attack_coeff
float release_coeff
limiter_state init_limiter(float attack_factor, float release_factor, int delay_len)
void limit(float *signal, int block_length, float threshold,
float *delay_line, limiter_state *state)
This is very similar to the C header file of the limiter:
typedef struct limiter_state_t {
int delay_index;
int delay_length;
float envelope;
float current_gain;
float attack_coeff;
float release_coeff;
} limiter_state;
limiter_state init_limiter(float attack_factor, float release_factor, int delay_len);
void limit(float *signal, int block_length, float threshold,
float *delay_line, limiter_state *state);
With that squared away, the C functions are accessible for Cython. Now, we only need a small Python wrapper around this code so it becomes usable from Python:
import numpy as np
cimport numpy as np
cimport limiter
DTYPE = np.float32
ctypedef np.float32_t DTYPE_t
cdef class Limiter:
cdef limiter.limiter_state state
cdef np.ndarray delay_line
def __init__(self, float attack_coeff, float release_coeff,
int delay_length):
self.state = limiter.init_limiter(attack_coeff, release_coeff, delay_length)
self.delay_line = np.zeros(delay_length, dtype=DTYPE)
def limit(self, np.ndarray[DTYPE_t,ndim=1] signal, float threshold):
limiter.limit(<float*>np.PyArray_DATA(signal),
<int>len(signal), threshold,
<float*>np.PyArray_DATA(self.delay_line),
<limiter.limiter_state*>&self.state)
The first two lines set this file up to access Numpy arrays both from the Python domain and the C domain, thus bridging the gap. The cimport limiter imports the C functions and types from above. The DTYPE stuff is advertising the Numpy float32 type to C.
The class is defined using cdef as a C data structure for speed. Also, Cython would naturally translate every C struct into a Python dict and vice versa, but we need to pass the struct to limit and have limit modify it. Thus, cdef limiter.limiter_state state makes Cython treat it as a C struct only. Finally, the np.PyArray_DATA() expressions expose the C arrays underlying the Numpy vectors. This is really handy since we don't have to copy any data around in order to modify the vectors from C.
As can be seen, the Cython implementation behaves nearly identically to the initial Python implementation (except for passing the dtype to the constructor) and can be used as a plug-in replacement (with the aforementioned caveat).
Finally, we need to build the whole contraption. The easiest way to do this is to use a setup file like this:
from distutils.core import setup
from distutils.extension import Extension
from Cython.Distutils import build_ext
from numpy import get_include
ext_modules = [Extension("cython_limiter",
sources=["cython_limiter.pyx",
"limiter.c"],
include_dirs=['.', get_include()])]
setup(
name = "cython_limiter",
cmdclass = {'build_ext': build_ext},
ext_modules = ext_modules
)
With that saved as setup.py, python setup.py build_ext --inplace will convert the Cython code to C, and then compile both the converted Cython code and C code into a binary Python module.
Conclusion
In this article, I developed a simple limiter and how to implement it in both C and Python. Then, I showed how to use the C implementation from Python. Where the Python implementation is struggling to keep a steady frame rate going even at large block sizes, the Cython version runs smoothly down to 2-4 samples per block on a 2 Ghz Core i7. Thus, real-time audio processing is clearly feasable using Python, Cython, Numpy and PyAudio.
You can find all the source code in this article at https://github.com/bastibe/simple-cython-limiter
Disclaimer
- I invented this limiter myself. I could invent a better sounding limiter, but this article is more about how to combine Python, Numpy, PyAudio and Cython for real-time signal processing than about limiter design.
- I recently worked on something similar at my day job. They agreed that I could write about it so long as I don't divulge any company secrets. This limiter is not a descendant of any code I worked on.
- Whoever wants to use any piece of code here, feel free to do so. I am hereby placing it in the public domain. Feel free to contact me if you have questions.
Fixing Errors in Epydoc
I ran into this error twice now and wasted an hour both times, so it is time to put this on my universal scratchpad, i.e. this blog.
If you ever get this error when using epydoc:
UNEXPECTED ERROR:
'Text' object has no attribute 'data'
You are probably running a version of Python that is greater than the latest one that is supported by epydoc. This is because epydoc has not been updated since 2008 and Python 2.5.
Luckily, some fine folks on the internet have figured out how to fix these things.
Short answer: Find your site-packages directory:
from distutils.sysconfig import get_python_lib print(get_python_lib())
Go there, navigate to the epydoc\/markup directory and change line 307 of the file restructuredtext.py from
m = self._SUMMARY_RE.match(child.data)
to
try: m = self._SUMMARY_RE.match(child.data) except AttributeError: m = None
This should fix that problem.
Compiling Scipy and Matplotlib again
Well, it's compile time again. Once again, I need to install scipy and matplotlib using homebrew and pip on Lion.
It seems things have improved since I tried to compile last time! Well, it still does not work out of the box, but at least now it can be done without compiling by hand:
(remember to brew install pkg-config gfortran first)
pip install -e git+https://github.com/scipy/scipy#egg=scipy-dev pip install -e git+https://github.com/matplotlib/matplotlib#egg=matplotlib
I must say, this is still a mess. But at least, it is getting less bad.
Compiling Scipy and Matplotlib using pip on Lion
So I upgraded to Lion. Predictably, some things went wrong. This time, the main thing that bit me was that for some reason, pip stopped working. After a bit of messing around with brew, pip and easy_install, I found out it was almost entirely my own fault. I messed up my PATH.
In the meantime, I had uninstalled all of brew's Python, so I had to reinstall. For me, that entails Python, Numpy, Scipy and Matplotlib. Only this time, Scipy would not build. Some obscure error in some veclib_cabi_c.c would report errors. A quick round of googling reveals:
In order to get Scipy to compile, you need to insert #include <complex.h> in
./scipy/lib/blas/fblaswrap_veclib_c.c.src
./scipy/linalg/src/fblaswrap_veclib_c.c
./scipy/sparse/linalg/eigen/arpack/ARPACK/FWRAPPERS/veclib_cabi_c.c
That done, Scipy compiles perfectly fine.
But, that is not enough yet. As this blogpost outlines, Matplotlib is not currently compatible with libpng 1.5, which ships with Lion. Fortunately, this is already fixed in the most recent source on the Matplotlib repo, so you just have to checkout that:
pip install -e git+https://github.com/matplotlib/matplotlib.git#egg=matplotlib
By doing that, Matplotlib should install just fine.
Seriously though, these PyPi repos are in a very sorry state. Every time I install one of these packages, I have to jump through hoops and spend hours debugging packages that really should work right out of the box. After all, brew, rvm and gem can do it just fine. Why is pip such a horrible mess?
Installing Pygame using Homebrew
So I want to do audio development on the Mac without using Matlab. An alternative to Matlab is Python, or rather, Numpy, Scipy and Matplotlib. They are awesome for working with audio data. What they don't do however is playing back audio. There are several packages out there that would afford audio playback. If you are serious about this though, you not only want audio playback, you want asynchronous audio playback. That is, you want to send some audio data to the sound card and continue with your program without waiting for the audio to finish playing. This allows continuous audio playback of computer-generated sound.
Pygame is one package that allows this. (I will submit a patch to Pyaudio soon that will enable it there, too). There are pre-built binaries on the Pygame website that you can install easily. But then there would be no easy way to uninstall them, so what I would rather want is to install Pygame using package managers that allow easy updating and uninstallation. My tool of choice on the Mac is of course Homebrew.
Note that although I am mostly interested in audio playback, this post will detail the installation of all modules of Pygame, not just pygame.mixer.
Homebrew won't install Pygame, but it will install all the prerequisites for Pygame. So, let's do that.
brew install sdl, sdl_mixer, sdl_ttf, libpng, jpeg, sdl_image, portmidi
This will install most packages for you. Note that libpng is also available as a system library, so it is installed keg_only, that is, without linking it in your path. We will need to compile against it though, so the next step is
brew link libpng
Now there is still one package missing, smpeg. Sadly, smpeg does not install its headers, so you can't compile against it. To fix that, type
brew edit smpeg
and add the following line just above the two end at the end of the file
include.install Dir["*.h"]
Then save the file. (I submitted a bug to have this fixed, so you might not need to do this when you read this). Now you can install smpeg with the usual
brew install smpeg
and you will get the headers, too. Isn't Homebrew great?
Now that all the prerequisites are met, lets look at Pygame itself. This is rather more difficult, as it will not build properly against Homebrew libraries on its own. First, download the source package of Pygame from the [official website](http://www.pygame.org/download.shtml). Unpack it to some directory.
Now open a terminal and navigate to that directory. Me, I like [iTerm](http://iterm.sourceforge.net/), but Terminal.app will do just fine, too. In there, run python config.py to create an initial setup file.
At this point, the setup file is mostly useless since config.py failed to find any homebrew-installed library. It is also strangely garbled, so there is some manual labor to do. Open the file Setup (no extension) in your favourite text editor. After the first comment block, you will see a line that looks like this
SDL = -I/NEED_INC_PATH_FIX -L/NEED_LIB_PATH_FIX -lSDL
Obviously, this is lacking the paths to the SDL library. If you installed Homebrew to its default directory, this will be in /usr/local…. Hence, change this line to
SDL = -I/usr/local/include/SDL -L/usr/local/lib -lSDL
The next lines are strangely garbled. They say, for example
FONT = -lS -lD -lL -l_ -lt -lt -lf
Where they actually should say
FONT = -lSDL_ttf
Instead of having one -l and then the library name SDL_ttf, they put -l in front of every single letter of the name. This is strange, and certainly wrong. So, correct it for FONT, IMAGE, MIXER and SMPEG.
Note that I did not tell you to do this for PORTTIME, too. Actually, PORTTIME is already correctly linked in PORTMIDI, so you don't need that at all any more. Just delete or comment the PORTTIME line.
Now that all the dependencies are corrected, lets enable the features. A few lines further down, there will be a block of lines, where most lines begin with a # except for the ones beginning with _numericsurfarray… and _camera…, These are the different features of Pygame: The ones with the # are disabled, the other two are enabled.
With all the stuff we installed earlier, you can now enable all features (remove the # in front of imageext…, font…, mixer…, mixer_music…, _minericsndarray…, movie…, scrap… and pypm…).
Remember we disabled PORTTIME a while ago? Right, so we have to remove that dependency: In the line starting with pypm…, delete the part that says $(PORTTIME). Great. That was easy, right? Now save that file and go back to the Terminal.
We are now going to compile and install Pygame. The nice thing is, even though we are installing it manually, it will go in the right directories and it will be registered with pip or easy_install, so you can just invoke them if you want to uninstall it later by typing pip uninstall pygame. This is something I love about Python!
Alright, now without further ado, install Pygame by typing
python setup.py install
Great! That's it! Everything should work now!
Installing Python/Numpy/Scipy/Matplotlib on OSX
For numerical analysis and signal processing prototyping, you would use Matlab. However, Matlab has some downsides that might make it unsuitable for your project. It might be too expensive. You might be a snobbish programmer that can't stand less-than-elegant programming languages. I certainly am.
So, you look for alternatives. You could take Octave, which is free, but that would not solve that ugly-code issue. You could take any scripting language you fancy, but Ruby, Perl and Python are too slow to do serious number crunching.
Then, you stumble upon that Python package called Numpy, which seems to be nearly as fast as Matlab when it comes to matrix processing and linear algebra. You then discover SciPy, which would add all that signal processing prowess of Matlab (do quick transformations, random numbers, statistics) to your toolbox. Last but not least, you need plotting. That would be Matplotlib then, which provides quick plotting facilities in Python.
And the best thing is, these three systems work really well together. They seem to be the perfect replacement for Matlab that could even be superiour to it in many regards.
Next up, you need to install all that stuff. If you are like me, you naturally want to do all that on a Mac. Also, you kind of dislike all these installer-thingies, which install stuff to unknown places and are nigh impossible to uninstall or update cleanly. Even though, you could of course just go to the individual websites, download Python, Numpy, SciPy and Matplotlib, run them installers, and be done. You would save yourself a lot of trouble that way.
But since you allegedly are like me, you instead fire up brew and try to install all that stuff using that. Again, you could use MacPorts or Fink instead, but you probably had some bad experiences with them and you generally love the hackishness of Homebrew, so this is your natural first try.
So you set about this, you believe in packet managers and trust them to take care of every obstacle that might be lying in your way. First of all, install the latest developer tools from developer.apple.com. You might need to register (for free) to get them. Also, you need to install Homebrew.
To cut this short, here is what you need to get that Python running:
brew install python
This one should be obvious. At the time of writing, it will install Python 2.7.1. You could take Python 3, but matplotlib is not compatible to it, so you kind of have to stick with 2.7.1 instead.
You also need to put /usr/local/bin and /usr/local/sbin in the beginning of your path to make sure the new Python gets loaded instead of the pre-installed one. You do that by writing
export PATH=/usr/local/bin:/usr/local/sbin:$PATH
in your \~/.bash_profile. (Create it if its not there–it is just a simple text file).
Now, if you type python --version, you should get Python 2.7.1 as a response.
Alright, next up, install the python package manager:
brew install distribute
brew install pip
This will come preconfigured for your newly installed Python. In an ideal world, this should be all. The world being as it is, the pip package of Matplotlib is severely broken and has one other unstated dependency:
brew install pkg-config
Also, SciPy is using some FORTRAN sources, so you need a fortran compiler:
brew install gfortran
Alright. That was enough. Now on to pip. With all these dependencies cleared, pip should be able to download Numpy and Scipy without trouble:
pip install numpy
pip install scipy
Matplotlib, on the other hand, is more difficult to install. You see, pip is looking at the Python package repository PyPi for each package. PyPi then provides a URL. Pip then scans that website for links to suitable package files. But, Sourceforge changed its links a while ago, so pip gets confused and will download an outdated version. Sourceforge says, its new links are way better and no way we will change them back; Pip says, well, if Sourceforge can't provide proper links, that's not our problem. Oh My. Silly children.
So we have to do this manually:
pip install -f http://sourceforge.net/projects/matplotlib/files/matplotlib/matplotlib-1.0.1/matplotlib-1.0.1.tar.gz matplotlib
That URL comes straight from Sourceforge. Look for the latest version of Matplotlib, search for the download link to the source distribution (*.tar.gz), copy that link and strip any trailing '/download'.
UPDATE:
It seems the matplotlib package was updated in the meantime, so you can just run pip install matplotlib now.
This should now download and install matplotlib.
Thank you for reading.
