Posts tagged "matlab":
Appending to Matlab Arrays
The variable $var appears to change size on every loop iteration. Consider preallocating for speed.
So sayeth Matlab.
Let's try it:
x_prealloc = cell(10000, 1);
x_end = {};
x_append = {};
for n=1:10000
% variant 1: preallocate
x_prealloc(n) = {42};
% variant 2: end+1
x_end(end+1) = {42};
% variant 3: append
x_append = [x_append {42}];
end
Which variant do you think is fastest?

Unsurprisingly, preallocation is indeed faster than growing an array. What is surprising is that it is faster by a constant factor of about 2 instead of scaling with the array length. Only appending by x = [x {42}] actually becomes slower for larger arrays. (The same thing happens for numerical arrays, struct arrays, and object arrays.)
TL;DR: Do not use x = [x $something], ever. Instead, use x(end+1) = $something. Preallocation is generally overrated.
Matlab Metaprogramming
Why is it, that I find Matlab to be a fine teaching tool and a fine tool for solving engineering problems, but at the same time, extremely cumbersome for my own work? Recently, the answer struck me: metaprogramming in Matlab sucks.
Matlab is marketed as a tool for engineers to solve engineering problems. There are convenient data structures for numerical data (arrays, tables), less convenient data structures for non-numeric data (cells, structs, chars), and a host of expensive but powerful functions and methods for working with this kind of data. This is the happy path.
But don't stray too far from the happy path; horrors lurk where The Mathworks don't dare going. Basic stuff like talking to sockets or interacting with other programs is very cumbersome in Matlab, and sometimes even downright impossible for the lack of threads, pipes, and similar infrastructure. But this is common knowledge, and consistent with Matlab's goals as an engineering tool, not a general purpose programming language. These are first-order problems, and they are rarely insurmountable.
The more insidious problem is metaprogramming, i.e. when the objects of your code are code objects themselves. The first order use of programming is to solve real-world problems. If these problems are numeric in nature, Matlab has got you covered. But as every programmer discovers at some point, the second order use of programming is to solve programming problems. And by golly, will Matlab let you down when you try that!
As soon as you climb that ladder of abstraction, and the objects of your code become code objects themselves, you will enter weird country. You think exist will tell you whether a variable name is taken? Try calling it on a method. You think nargout will always give you a number? Again, methods will enlighten you. Quick, how do you capture all output arguments of a function call in a variable? x = cell(nargout(fun), 1); [x{:}] = fun(...), obviously (this sometimes fails). And don't even think of trying to overload subsref to create something generic. Those subsref semantics are crazy talk!
I could go on. The real power of code is abstraction. We use programming to repeatedly and reliably solve similar problems. The logical next step is to use those same programming tools to solve similar kinds of problems. This happens to all of us, and Matlab makes it extremely hard to deal with. Thus, it puts a ceiling on the level of abstraction that is reasonably achievable, and limits engineers to first-order solutions. And after a few years of acclimatization, it will put that same ceiling on those engineers' thinking, because you can't reason about what you can't express.
MATLAB Syntax
In a recent project, I tried to parse MATLAB code. During this trying exercise, I stumbled upon a few… unique design decisions of the MATLAB language:
Use of apostrophes (')
Apostrophes can mean one of two things: If applied as a unary postfix operator, it means transpose. If used as a unary prefix operator, it marks the start of a string. While not a big problem for human readers, this makes code surprisingly hard to parse. The interesting bit about this, though, is the fact that there would have been a much easier way to do this: Why not use double quotation marks for strings, and apostrophes for transpose? The double quotation mark is never used in MATLAB, so this would have been a very easy choice.
Use of parens (())
If Parens follow a variable name, they can mean one of two things: If the variable is a function, the parens denote a function call. If the variable is anything else, this is an indexing operation. This can actually be very confusing to readers, since it makes it entirely unclear what kind of operation foo(5) will execute without knowledge about foo (which might not be available until runtime). Again, this could have been easily solved by using brackets ([]) for indexing, and parens (()) for function calls.
Use of braces ({}) and cell arrays
Cell arrays are multi-dimensional, ordered, heterogeneous collections of things. But in contrast to every other collection (structs, objects, maps, tables, matrices), they are not indexed using parens, but braces. Why? I don't know. In fact, you can index cell arrays using parens, but this only yields a new cell array with only one value. Why would this ever be useful? I have no explanation. This constantly leads to errors, and for the life of me I can not think of a reason for this behavior.
Use of line breaks
In MATLAB, your line can end on one of three characters: A newline character, a semicolon (;), and a comma (,). As we all know, the semicolon suppresses program output, while the newline character does not. The comma ends the logical line, but does not suppress program output. This is a relatively little-known feature, so I thought it would be useful to share it. Except, the meaning of ; and , changes in literals (like [1, 2; 3, 4] or {'a', 'b'; 3, 4}). Here, commas separate values on the same row and are optional, and semicolons end the current row. Interestingly, literals also change the meaning of the newline character: Inside a literal, a newline acts just like a semicolon, overrides a preceding comma, and you don't have to use ellipsis (...) for line continuations.
Syntax rules for commands
Commands are function calls without the parenthesis, like help disp, which is syntactically equivalent to help('disp'). You see, if you just specify a function name (can't be a compound expression or a function handle), and don't use parenthesis, all following words will be interpreted as strings, and passed to the function. This is actually kind of a neat feature. However, how do you differentiate between variable_name + 5 and help + 5? The answer is: Commands are actually a bit more complex. A command starts with a function name, followed by a space, which is not followed by an operator and a space. Thus, help +5 + 4 is a command, while help + 5 + 4 is an addition. Tricky!
The more-than-one-value value
If you want to save more than one value in a variable, you can use a collection (structs, matrices, maps, tables, cell arrays). In addition though, MATLAB knows another way of handling more than one values at once: The thing you get when you index a cell array with {:} or assign a function call with more than one result. In that case, you get something that is assignable to several variables, but that is not itself a collection. Just another quirk of MATLAB's indexing logic. However, you can capture these values into matrices or cell arrays using brackets or braces, like this: {x{:}} or [x{:}]. Note that this also works in assignments in a confusing way: [z{:}] = x{:} (if both x and z have the same length). Incidentally, this is often a neat way of converting between different kinds of collections (but utterly unreadable, because type information is hopelessly lost).
Transplant, revisited
A few months ago, I talked about the performance of calling Matlab from Python. Since then, I implemented a few optimizations that make working with Transplant a lot faster:
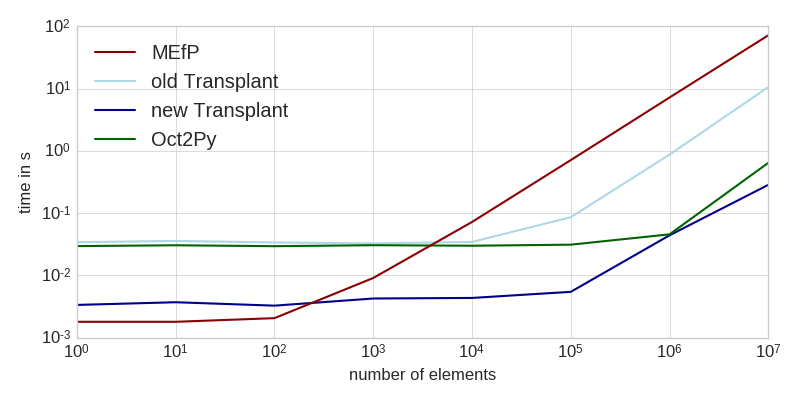
The workload consisted of generating a bunch of random numbers (not included in the times), and sending them to Matlab for computation. This task is entirely dominated by the time it takes to transfer the data to Matlab (see table at the end for intra-language benchmarks of the same task).
As you can see, the new Transplant is significantly faster for small workloads, and still a factor of two faster for larger amounts of data. It is now almost always a faster solution than the Matlab Engine for Python (MEfP) and Oct2Py. For very large datasets, Oct2Py might be preferable, though.
This improvement comes from three major changes: Matlab functions are now returned as callable objects instead of ad-hoc functions, Transplant now uses MsgPack instead of JSON, and loadlibrary instead of a Mex file to call into libzmq. All of these changes are entirely under the hood, though, and the public API remains unchanged.
The callable object thing is the big one for small workloads. The advantage is that the objects will only fetch documentation if __doc__ is actually asked for. As it turns out, running help('funcname') for every function call is kind of a big overhead.
Bigger workloads however are dominated by the time it takes Matlab to decode the data. String parsing is very slow in Matlab, which is a bad thing indeed if you're planning to read a couple hundred megabytes of JSON. Thus, I replaced JSON with MsgPack, which eliminates the parsing overhead almost entirely. JSON messaging is still available, though, if you pass msgformat='json' to the constructor. Edit: Additionally, binary data is no longer encoded as base64 strings, but passed directly through MsgPack. This yields about a ten-fold performance improvement, especially for larger data sets.
Lastly, I rewrote the ZeroMQ interaction to use loadlibrary instead of a Mex file. This has no impact on processing speed at all, but you don't have to worry about compiling that C code any more.
Oh, and Transplant now works on Windows!
Here is the above data again in tabular form:
| Task | New Transplant | Old Transplant | Oct2Py | MEfP | Matlab | Numpy | Octave |
|---|---|---|---|---|---|---|---|
| startup | 4.8 s | 5.8 s | 11 ms | 4.6 s | |||
| sum(randn(1,1)) | 3.36 ms | 34.2 ms | 29.6 ms | 1.8 ms | 9.6 μs | 1.8 μs | 6 μs |
| sum(randn(1,10)) | 3.71 ms | 35.8 ms | 30.5 ms | 1.8 ms | 1.8 μs | 1.8 μs | 9 μs |
| sum(randn(1,100)) | 3.27 ms | 33.9 ms | 29.5 ms | 2.06 ms | 2.2 μs | 1.8 μs | 9 μs |
| sum(randn(1,1000)) | 4.26 ms | 32.7 ms | 30.6 ms | 9.1 ms | 4.1 μs | 2.3 μs | 12 μs |
| sum(randn(1,1e4)) | 4.35 ms | 34.5 ms | 30 ms | 72.2 ms | 25 μs | 5.8 μs | 38 μs |
| sum(randn(1,1e5)) | 5.45 ms | 86.1 ms | 31.2 ms | 712 ms | 55 μs | 38.6 μs | 280 μs |
| sum(randn(1,1e6)) | 44.1 ms | 874 ms | 45.7 ms | 7.21 s | 430 μs | 355 μs | 2.2 ms |
| sum(randn(1,1e7)) | 285 ms | 10.6 s | 643 ms | 72 s | 3.5 ms | 5.04 ms | 22 ms |
Teaching with Matlab Live Scripts
For a few years now, I have been teaching programming courses using notebooks. A notebook is an interactive document that can contain code, results, graphs, math, and prose. It is the perfect teaching tool:
You can combine introductory resources with application examples, assignments, and results. And after the lecture, students can refer to these notebooks at their leisure, and re-run example code, or try different approaches with known data.
The first time I saw this was with the Jupyter notebook (née IPython notebook). I immediately used it for teaching an introductory programming course in Python.
Later, I took over a Matlab course, but Matlab lacked a notebook. So for the next two years of teaching Matlab, I hacked up a small IPython extension that allowed me to run Matlab code in an Jupyter notebook as a cell magic.
Now, with 2016a, Matlab introduced Live Scripts, which is Mathworkian for notebook. This blog post is about how Live Scripts compare to Jupyter notebooks.
First off, Live Scripts work. The basic functionality is there: Code, prose, figures, and math can be saved in one document; The notebook can be exported as PDF and HTML, and Students can download the notebook and play with it. This latter part was not possible with my homegrown solution earlier.
However, Live Scripts are new, and still contain a number of bugs. You can't customize figure sizes, formatting options are very basic, image rendering is terrible, and math rendering using LaTeX is of poor quality and limited. Also, using Live Scripts on a retina Mac is borderline impossible: Matlab crashes on screen resolution changes (i.e. connecting a projector), Live Scripts render REALLY slowly (type a word, watch the characters crawl onto the screen one by one), and all figures export in twice their intended size (fixed in 2016b). You can work around some of these issues by starting Matlab in Low Resolution Mode.
No doubt some of these issues are going to get addressed in future releases. 2016b added script-local functions, which I read mostly as "you can now write functions in Live Scripts", and autocorrection-like text replacements that convert Markdown formatting into formatted text. This is highly appreciated.
Additionally though, here are a few features I would love to see:
- Nested lists, and lists entries that contain newlines (i.e. differentiate between line breaks and paragraph breaks).
- Indented text, for quoting things, or to work around the lack of multi-line list entries.
- More headline levels.
- Magics. This is probably a long shot, but line/cell magics in Jupyter notebooks are really useful.
Still, all griping aside, I want to reiterate that Live Scripts work. They aren't quite as nice as Jupyter notebooks, but they serve their purpose, and are a tremendously useful teaching tool.
Matlab has an FFI and it is not Mex
Sometimes, you just have to use C code. There's no way around it. C is the lingua franca and bedrock of our computational world. Even in Matlab, sometimes, you just have to call into a C library.
So, you grab your towel, you bite the bullet, you strap into your K&R, and get down to it: You start writing a Mex file. And you curse, and you cry, because writing C is hard, and Mex doesn't exactly make it any better. But you know what? There is a better way!
Because, unbeknownst to many, Matlab includes a Foreign Function Interface. The technique was probably pioneered by Common Lisp, and has since been widely adopted everywhere: calling functions in a C library without writing any C code and without invoking a compiler!
Mind you, there remains a large and essential impedance mismatch between C's statically typed calling conventions and the vagaries of a dynamically typed language such as Matlab, and even the nicest FFI can't completely hide that fact. But anything is better than the abomination that is Mex.
So here goes, a very simple C library that adds two arrays:
// test.c:
#include test.h
void add_arrays(float *out, float* in, size_t length) {
for (size_t n=0; n<length; n++) {
out[n] += in[n];
}
}
// test.h:
#include <stddef.h>
void add_arrays(float *out, float *in, size_t length);
Let's compile it! gcc -shared -std=c99 -o test.so test.c will do the trick.
Now, let's load that library into Matlab with loadlibrary:
if not(libisloaded('test'))
[notfound, warnings] = loadlibrary('test.so', 'test.h');
assert(isempty(notfound), 'could not load test library')
end
Note that loadlibrary can't parse many things you would commonly find in header files, so you will likely have to strip them down to the bare essentials. Additionally, loadlibrary doesn't throw errors if it can't load a library, so we always have to check the notfound output argument to see if the library was actually loaded successfully.
With that, we can call functions in that library using calllib. But we can't just pass in Matlab vectors, that would be too easy. We first have to convert them to something C can understand: Pointers
vector1 = [1 2 3 4 5];
vector2 = [9 8 7 6 5];
vector1ptr = libpointer('singlePointer', vector1);
vector2ptr = libpointer('singlePointer', vector2);
What is nice about this is that this automatically converts the vectors from double to float. What is less nice is that it uses its weird singlePtr notation instead of the more canonical float* that you would expect from a self-respecting C header.
Then, finally, let's call our function:
calllib('test', 'add_arrays', vector1ptr, vector2ptr, length(vector1));
If you see no errors, everything went smoothly, and you will now have changed the content of vector1ptr, which we can have a look at like this:
added_vectors = vector1ptr.Value;
Note that this didn't change the contents of vector1, only of the newly created pointer. So there will always be some memory overhead to this technique in comparison to Mex files. However, runtime overhead seems pretty fine:
timeit(@() calllib('test', 'add_arrays', vector1ptr, vector2ptr, length(vector1)))
% ans = 1.9155e-05
timeit(@() the_same_thing_but_as_a_mex_file(single(vector1), single(vector2)))
% ans = 4.6262e-05
timeit(@() the_same_thing_plus_argument_conversion(vector1, vector2))
% ans = 1.2326e-04
So as you can see, the calllib is plenty fast. However, if you add the Matlab code for converting the double arrays to pointers and extracting the summed data afterwards, the FFI is noticeably slower than a Mex file.
However, If I ask myself whether I would sacrifice 0.00007 seconds of computational overhead for hours of my life not spent with Mex, there really is no competition. I will choose Matlab's FFI over writing Mex files every time.
Calling Matlab from Python
For my latest experiments, I needed to run both Python functions and Matlab functions as part of the same program. As I noted earlier, Matlab includes the Matlab Engine for Python (MEfP), which can call Matlab functions from Python. Before I knew about this, I created Transplant, which does the very same thing. So, how do they compare?
Usage
As it's name suggests, Matlab is a matrix laboratory, and matrices are the most important data type in Matlab. Since matrices don't exist in plain Python, the MEfP implements it's own as matlab.double et al., and you have to convert any data you want to pass to Matlab into one of those. In contrast, Transplant recognizes the fact that Python does in fact know a really good matrix engine called Numpy, and just uses that instead.
Matlab Engine for Python | Transplant
---------------------------------------|---------------------------------------
import numpy | import numpy
import matlab | import transplant
import matlab.engine |
|
eng = matlab.engine.start_matlab() | eng = transplant.Matlab()
numpy_data = numpy.random.randn(100) | numpy_data = numpy.random.randn(100)
list_data = numpy_data.tolist() |
matlab_data = matlab.double(list_data) |
data_sum = eng.sum(matlab_data) | data_sum = eng.sum(numpy_data)
Aside from this difference, both libraries work almost identical. Even the handling of the number of output arguments is (accidentally) almost the same:
Matlab Engine for Python | Transplant
---------------------------------------|---------------------------------------
eng.max(matlab_data) | eng.max(numpy_data)
>>> 4.533 | >>> [4.533 537635]
eng.max(matlab_data, nargout=1) | eng.max(numpy_data, nargout=1)
>>> 4.533 | >>> 4.533
eng.max(matlab_data, nargout=2) | eng.max(numpy_data, nargout=2)
>>> (4.533, 537635.0) | >>> [4.533 537635]
Similarly, both libraries can interact with Matlab objects in Python, although the MEfP can't access object properties:
Matlab Engine for Python | Transplant
---------------------------------------|---------------------------------------
f = eng.figure() | f = eng.figure()
eng.get(f, 'Position') | eng.get(f, 'Position')
>>> matlab.double([[ ... ]]) | >>> array([[ ... ]])
f.Position | f.Position
>>> AttributeError | >>> array([[ ... ]])
There are a few small differences, though:
- Function documentation in the MEfP is only available as
eng.help('funcname'). Transplant will populate a function's__doc__, and thus documentation tools like IPython's?operator just work. - Transplant converts empty matrices to
None, whereas the MEfP represents them asmatlab.double([]). - Transplant represents
dictascontainers.Map, while the MEfP usesstruct(the former is more correct, the latter arguable more useful). - If the MEfP does not know
nargout, it assumesnargout=1. Transplant usesnargout(func)or returns whatever the function writes intoans. - The MEfP can't return non-scalar structs, such as the return value of
whos. Transplant can do this. - The MEfP can't return anonymous functions, such as
eng.eval('@(x, y) x>y'). Transplant can do this.
Performance
The time to start a Matlab instance is shorter in MEfP (3.8 s) than in Transplant (6.1 s). But since you're doing this relatively seldomly, the difference typically doesn't matter too much.
More interesting is the time it takes to call a Matlab function from Python. Have a look:

This is running sum(randn(n,1)) from Transplant, the MEfP, and in Matlab itself. As you can see, the MEfP is a constant factor of about 1000 slower than Matlab. Transplant is a constant factor of about 100 slower than Matlab, but always takes at least 0.05 s.
There is a gap of about a factor of 10 between Transplant and the MEfP. In practice, this gap is highly significant! In my particular use case, I have a function that takes about one second of computation time for an audio signal of ten seconds (half a million values). When I call this function with Transplant, it takes about 1.3 seconds. With MEfP, it takes 4.5 seconds.
Transplant spends its time serializing the arguments to JSON, sending that JSON over ZeroMQ to Matlab, and parsing the JSON there. Well, to be honest, only the parsing part takes any significant time, overall. While it might seem onerous to serialize everything to JSON, this architecture allows Transplant to run over a network connection.
It is a bit baffling to me that MEfP manages to be slower than that, despite being written in C. Looking at the number of function calls in the profiler, the MEfP calls 25 functions (!) on each value (!!) of the input data. This is a shockingly inefficient way of doing things.
TL;DR
It used to be very difficult to work in a mixed-language environment, particularly with one of those languages being Matlab. Nowadays, this has thankfully gotten much easier. Even Mathworks themselves have stepped up their game, and can interact with Python, C, Java, and FORTRAN. But their interface to Python does leave something to be desired, and there are better alternatives available.
If you want to try Transplant, just head over to Github and use it. If you find any bugs, feature requests, or improvements, please let me know in the Github issues.
Massive Memory Leak in the Matlab Engine for Python
As of Matlab 2014b, Matlab includes a Python module for calling Matlab code from Python. This is how you use it:
import numpy import matlab import matlab.engine eng = matlab.engine.start_matlab() random_data = numpy.random.randn(100) # convert Numpy data to Matlab: matlab_data = matlab.double(random_data.tolist()) data_sum = eng.sum(matlab_data)
You can call any Matlab function on eng, and you can access any Matlab workspace variable in eng.workspace. As you can see, the Matlab Engine is not Numpy-aware, and you have to convert all your Numpy data to Matlab double before you can call Matlab functions with it. Still, it works pretty well.
Recently, I ran a rather large experiment set, where I had a set of four functions, two in Matlab, two in Python, and called each of these functions a few thousand times with a bunch of different data to see how they performed.
While doing that I noticed that my Python processes were growing larger and larger, until they consumed all my memory and a sizeable chunk of my swap as well. I couldn't find any reason for this. None of my Python code cached anything, and the sum total of all global variables did not amount to anything substantial.
Enter Pympler, a memory analyzer for Python. Pympler is an amazing library for introspecting your program's memory. Among its many features, it can list the biggest objects in your running program:
from pympler import muppy, summary summary.print_(summary.summarize(muppy.get_objects()))
types | # objects | total size
=========================================== | =========== | ============
<class 'array.array | 1076 | 2.77 GB
<class 'str | 42839 | 7.65 MB
<class 'dict | 8604 | 5.43 MB
<class 'numpy.ndarray | 48 | 3.16 MB
<class 'code | 14113 | 1.94 MB
<class 'type | 1557 | 1.62 MB
<class 'list | 3158 | 1.38 MB
<class 'set | 1265 | 529.72 KB
<class 'tuple | 5129 | 336.98 KB
<class 'bytes | 2413 | 219.48 KB
<class 'weakref | 2654 | 207.34 KB
<class 'collections.OrderedDict | 65 | 149.85 KB
<class 'wrapper_descriptor | 1676 | 130.94 KB
<class 'traitlets.traitlets.MetaHasTraits | 107 | 123.55 KB
<class 'getset_descriptor | 1738 | 122.20 KB
Now that is interesting. Apparently, I was lugging around close to three gigabytes worth of bare-Python array.array. And these are clearly not Numpy arrays, since those would show up as numpy.ndarray. But I couldn't find any of these objects in my workspace.
So let's get a reference to one of these objects, and see who they belong to. This can also be done with Pympler, but I prefer the way objgraph does it:
import array # get a list of all objects known to Python: all_objects = muppy.get_objects() # sort out only `array.array` instances: all_arrays = [obj for obj in all_objects if isinstance(obj, array.array)] import objgraph objgraph.show_backrefs(all_arrays[0], filename='array.png')
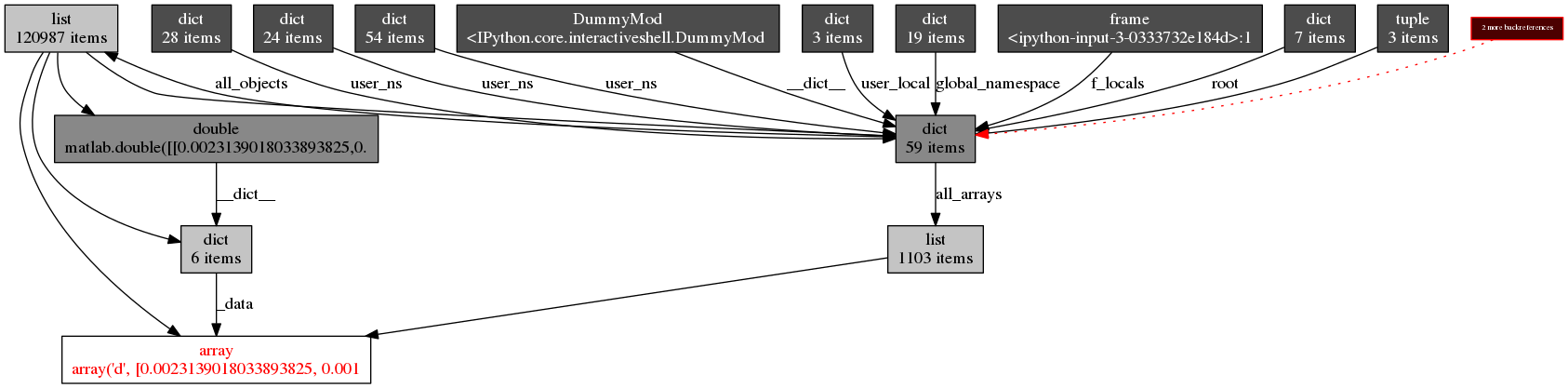
It seems that the array.array object is part of a matlab.double instance which is not referenced from anywhere but all_objects. A memory leak.
After a bit of experimentation, I found the culprit. To illustrate, here's an example: The function leak passes some data to Matlab, and calculates a float. Since the variables are not used outside of leak, and the function does not return anything, all variables within the function should get deallocated when leak returns.
def leak():
test_data = numpy.zeros(1024*1024)
matlab_data = matlab.double(test_data.tolist())
eng.sum(matlab_data)
Pympler has another great feature that can track allocations. The SummaryTracker will track and display any allocations between calls to print_diff(). This is very useful to see how much memory was used during the call to leak:
from pympler import tracker tr = tracker.SummaryTracker() tr.print_diff() leak() tr.print_diff()
types | # objects | total size
========================== | =========== | ============
<class 'array.array | 1 | 8.00 MB
...
And there you have it. Note that this leak is not the Numpy array test_data and it is not the matlab array matlab_data. Both of these are garbage collected correctly. But the Matlab Engine for Python will leak any data you pass to a Matlab function.
This data is not referenced from anywhere within Python, and is counted as leaked by objgraph. In other words, the C code inside the Matlab Engine for Python copies all passed data into it's internal memory, but never frees that memory. Not even if you quit the Matlab Engine, or del all Python references to it. Your only option is to restart Python.
Postscriptum
I since posted a bug report on Mathworks, and received a patch that fixes the problem. Additionally, Mathworks said that the problem only occurs on Linux.
Matlab and Audio Files
So I wanted to work with audio files in Matlab. In the past, Matlab could only do this with auread and wavread, which can read *.au and *.wav files. With 2012b, Matlab introduced audioread, which claims to support *.wav, *.ogg, *.flac, *.au, *.mp3, and *.mp4, and simultaneously deprecated auread and wavread.
Of these file formats, only *.au is capable of storing more than 4 Gb of audio data. But the documentation is actually wrong: audioread can actually read more data formats than documented: it reads *.w64, *.rf64, and *.caf no problem. And these can store more than 4 Gb as well.
It's just that, while audioread supports all of these nice file formats, audiowrite is more limited, and only supports *.wav, *.ogg, *.flac, and *.mp4. And it does not support any undocumented formats, either. So it seems that there is no way of writing files larger than 4 Gb. But for the time being, auwrite is still available, even though deprecated. I tried it, though, and it didn't finish writing 4.8 Gb in half an hour.
In other words, Matlab is incapable of writing audio files larger than 4 Gb. It just can't do it.
Unicode and Matlab on the command line
As per the latest Stackoverflow Developer Survey, Matlab is one of the most dreaded tools out there. I run into Matlab-related trouble daily. In all honesty, I have never seen a programming language as user-hostile and as badly designed as this.
So here is today's problem: When run from the command line, Matlab does not render unicode characters (on OSX).
I say "(on OSX)", because on Windows, it does not print a damn thing. Nope, no disp output for Windows users.
More analysis: It's not that Matlab does not render unicode characters at all when run from the command line. Instead, it renders them as 0x1a aka SUB aka substitute character. In other words, it tries to render unicode as ASCII (which doesn't work), and then replaces all non-ASCII characters with SUB. This is actually reasonable if Matlab were running on a machine that can't handle unicode. This is not a correct assessment of post-90s Macs, though.
To see why Matlab would do such a dastardly deed, you can use feature('locale') to get information about the encoding Matlab uses. On Windows and OS X, this defaults to either ISO-8859-1 (when your locale is pure de_DE or en_US) or US-ASCII, if it is something impure. In my case, German dates but English text. Because US-ASCII is obviously the most all-encompassing choice for such mixed-languages environments.
But luckily, there is help. Matlab has a widely documented (not) and easily discoverable (not) configuration option to change this: To change Matlab's encoding settings, edit %MATLABROOT%/bin/lcdata.xml, and look for the entry for your locale. For me, this is one of
<locale name="de_DE" encoding="ISO-8859-1" xpg_name="de_DE.ISO8859-1"> ... <locale name="en_US" encoding="ISO-8859-1" xpg_name="en_US.ISO8859-1"> ...
In order to make Matlab's encoding default to UTF-8, change the entry for your locale to
<locale name="de_DE" encoding="UTF-8" xpg_name="de_DE.UTF-8"> ... <locale name="en_US" encoding="UTF-8" xpg_name="en_US.UTF-8"> ...
With that, Matlab will print UTF-8 to the terminal.
You still can't type unicode characters to the command prompt, of course. But who would want that anyway, I dare ask. Of course, what with Matlab being basically free, and frequently updated, we can forgive such foibles easily…
Transplant
In academia, a lot of programming is done in Matlab. Many very interesting algorithms are only available in Matlab. Personally, I prefer to use tools that are more widely applicable, and less proprietary, than Matlab. My weapon of choice at the moment is Python.
But I still need to use Matlab code. There are a few ways of interacting with Matlab out there already. Most of them focus on being able to eval strings in Matlab. Boring. The most interesting one is mlab, a full-fledget bridge between Python and Matlab! Had I found this earlier, I would probably not have written my own.
But write my own I did: Transplant. Transplant is a very simple bridge for calling Matlab functions from Python. Here is how you start Matlab from Python:
import transplant matlab = transplant.Matlab()
This matlab object starts a Matlab interpreter in the background and connects to it. You can call Matlab functions on it!
matlab.eye(3)
>>> array([[ 1., 0., 0.],
[ 0., 1., 0.],
[ 0., 0., 1.]])
As you can see, Matlab matrices are converted to Numpy matrices. In contrast to most other Python/Matlab bridges, matrix types are preserved1:
matlab.randi(255, 1, 4, 'uint8') >>> array([[246, 2, 198, 209]], dtype=uint8)
All matrix data is actually transferred in binary, so both Matlab and Python work on bit-identical data. This is very important if you are working with precise data! Most other bridges do some amount of type conversion at this point.
This alone accounts for a large percentage of Matlab code out there. But not every Matlab function can be called this easily from Python: Matlab functions behave differently depending the number of output arguments! To emulate this in Python, every function has a keyword argument nargout 2. For example, the Matlab function max by default returns both the maximum value and the index of that value. If given nargout=1 it will only return the maximum value:
data = matlab.randn(1, 4) matlab.max(data) >>> [1.5326, 3] # Matlab: x, n = max(...) matlab.max(data, nargout=1) >>> 1.5326 # Matlab: x = max(...)
If no nargout is given, functions behave according to nargout(@function). If even that fails, they return the content of ans after their execution.
Calling Matlab functions is the most important feature of Transplant. But there is a more:
You can save/retrieve variables in the global workspace:
matlab.value = 5 # Matlab: value = 5 x = matlab.value # Matlab: x = value
You van eval some code:
matlab.eval('class(value)') >>> ans = >>> >>> double >>>- The help text for functions is automatically assigned as docstring. In IPython, this means that
matlab.magic?displays the same thinghelp magicwould display in Matlab.
Under the hood, Transplant is using a very simple messaging protocol based on 0MQ, JSON, and some base64-encoded binary data. Sadly, Matlab can deal with none of these technologies by itself. Transplant therefore contains a full-featured JSON parser/serializer and base64 encoder/decoder in pure Matlab. It also contains a minimal mex-file for interfacing with 0MQ.
There are a few JSON parsers available for Matlab, but virtually all of them try parse JSON arrays as matrices. This means that these parsers have no way of differentiating between a list of vectors and a matrix (want to call a function with three vectors or a matrix? No can do). Transplant's JSON parser parses JSON arrays as cell arrays and JSON objects as structs. While somewhat less convenient in general, this is a much better fit for transferring data structures between programming languages.
Similarly, there are a few base64 encoders available. Most of them actually use Matlab's built-in Java interface to encode/decode base64 strings. I tried this, but it has two downsides: Firstly, it is pretty slow for short strings since the data has to be copied over to the Java side and then back. Secondly, it is limited by the Java heap space. I was not able to reliably encode/decode more than about 64 Mb using this3. My base64 encoder/decoder is written in pure Matlab, and works for arbitrarily large data.
All of this has been about Matlab, but my actual goal is bigger: I want transplant to become a library for interacting between more than just Python and Matlab. In particular, Julia and PyPy would be very interesting targets. Also, it would be useful to reverse roles and call Python from Matlab as well! But that will be in the future.
For now, head over to Github.com/bastibe/transplant and have fun! Also, if you find any bugs or have any suggestions, please open an issue on Github!
Footnotes:
A Python Primer for Matlab Users
Why would you want to use Python over Matlab?
- Because Python is free and Matlab is not.
- Because Python is a general purpose programming language and Matlab is not.
Let me qualify that a bit. Matlab is a very useful programming environment for numerical problems. For a very particular set of problems, Matlab is an awesome tool. For many other problems however, it is just about unusable. For example, you would not write a complex GUI program in Matlab, you would not write your blogging engine in Matlab and you would not write a web service in Matlab. You can do all that and more in Python.
Python as a Matlab replacement
The biggest strength of Matlab is its matrix engine. Most of the data you work with in Matlab are matrices and there is a host of functions available to manipulate and visualize those matrices. Python, by itself, does not have a convenient matrix engine. However, there are three packages (think Matlab Toolboxes) out there that will add this capability to Python:
- Numpy (the matrix engine)
- Scipy (matrix manipulation)
- Matplotlib (plotting)
You can either grab the individual installers for Python, Numpy, Scipy and Matplotlib from their respective websites, or get them pre-packaged from pythonxy() or EPD.
A 30,000 foot overview
Like Matlab, Python is interpreted, that is, there is no need for a compiler and code can be executed at any time as long as Python is installed on the machine. Also, code can be copied from one machine to another and will run without change.
Like Matlab, Python is dynamically typed, that is, every variable can hold data of any type, as in:
# Python
a = 5 # a number
a = [1, 2, 3] # a list
a = 'text' # a string
Contrast this with C, where you can not assign different data types to the same variable:
// C
int a = 5;
float b[3] = {1.0, 2.0, 3.0};
char c[] = "text";
Unlike Matlab, Python is strongly typed, that is, you can not add a number to a string. In Matlab, adding a single number to a string will convert that string into an array of numbers, then add the single number to each of the numbers in the array. Python will simply throw an error.
% Matlab
a = 'text'
b = a + 5 % [121 106 125 121]
# Python
a = 'text'
b = a + 5 # TypeError: Can't convert 'int' object to str implicitly
Unlike Matlab, every Python file can contain as many functions as you like. Basically, you can organize your code in as many files as you want. To access functions from other files, use import filename.
Unlike Matlab, Python is very quick to start. In fact, most operating systems automatically start a new Python process whenever you run a Python program and quit that process once the program has finished. Thus, every Python program behaves as if it indeed were an independent program. There is no need to wait for that big Matlab mother ship to start before writing or executing code.
Unlike Matlab, the source code of Python is readily available. Every detail of Python's inner workings is available to everyone. It is thus feasible and encouraged to actively participate in the development of Python itself or some add-on package. Furthermore, there is no dependence on some company deciding where to go next with Python.
Reading Python
When you start up Python, it is a rather empty environment. In order to do anything useful, you first have to import some functionality into your workspace. Thus, you will see a few lines of import statements at the top of every Python file. Moreover, Python has namespaces, so if you import numpy, you will have to prefix every feature of Numpy with its name, like this:
import numpy
a = numpy.zeros(10, 1)
This is clearly cumbersome if you are planning to use Numpy all the time. So instead, you can import all of Numpy into the global environment like this:
from numpy import *
a = ones(30, 1)
Better yet, there is a pre-packaged namespace that contains the whole Numpy-Scipy-Matplotlib stack in one piece:
from pylab import *
a = randn(100, 1)
plot(a)
show()
Note that Python does not plot immediately when you type plot(). Instead, it will collect all plotting information and only show it on the screen once you type show().
So far, the code you have seen should look pretty familiar. A few differences:
- No semicolons at the end of lines;
In order to print stuff to the console, use the
print()function instead. - No
endanywhere. In Python, blocks of code are identified by indentation and they always start with a colon like so:
sum = 0
for n in [1, 2, 3, 4, 5]:
sum = sum + n
print(sum)
- Function definitions are different.
They use the
defkeyword instead offunction. You don't have to name the output variable names in the definition and instead usereturn().
# Python
def abs(number):
if number > 0:
return number
else:
return -number
% Matlab
function [out] = abs(number)
if number > 0
out = number
else
out = -number
end
end
- There is no easy way to write out a list or matrix.
Since Python only gains a matrix engine by importing Numpy, it does not have a convenient way of writing arrays or matrices. This sounds more inconvenient than it actually is, since you are probably using mostly functions like
zeros()orrandn()anyway and those work just fine. Also, many places accept Python lists (like this[1, 2, 3]) instead of Numpy arrays, so this rarely is a problem. Note that you must use commas to separate items and can not use semicolons to separate lines.
# create a numpy matrix:
m = array([[1, 2, 3],
[4, 5, 6],
[7, 8, 9]])
# create a Python list:
l = [1 2 3]
- Arrays access uses brackets and is numbered from 0.
Thus, ranges exclude the last number (see below).
Mostly, this just means that array access does not need any
+1or-1when indexing arrays anymore.
a = linspace(1, 10, 10)
one = a[0]
two = a[1]
# "6:8" is a range of two elements:
a[6:8] = [70, 80] # <-- a Python list!
Common traps
- Array slicing does not copy.
a = array([1 2 3 4 5])
b = a[1:4] # [2 3 4]
b[1] = rand() # this will change a and b!
# make a copy like this:
c = array(a[1:4], copy=True) # copy=True can be omitted
c[1] = rand() # changes only c
- Arrays retain their data type.
You can slice them, you can dice them, you can do math on them, but a 16 bit integer array will never lose its data type. Use
new = array(old, dtype=double)to convert an array of any data type to the defaultdoubletype (like in Matlab).
# pretend this came from a wave file:
a = array([1000, 2000, 3000, 4000, 5000], dtype=int16)
a = a * 10 # int16 only goes to 32768!
# a is now [10000, 20000, 30000, -25536, -15536]
Going further
Now you should be able to read Python code reasonably well. Numpy, Scipy and Matplotlib are actually modeled after Matlab in many ways, so many functions will have a very similar name and functionality. A lot of the numerical code you write in Python will look very similar to the equivalent code in Matlab. For a more in-depth comparison of Matlab and Python syntax, head over to the Numpy documentation for Matlab users.
However, since Python is a general purpose programming language, it offers some more tools. To begin with, there are a few more data types like associative arrays, tuples (unchangeable lists), proper strings and a full-featured object system. Then, there is a plethora of add-on packages, most of which actually come with your standard installation of Python. For example, there are internet protocols, GUI programming frameworks, real-time audio interfaces, web frameworks and game development libraries. Even this very blog is created using a Python static site generator.
Lastly, Python has a great online documentation site including a tutorial, there are many books on Python and there is a helpful Wiki on Python. There is also a tutorial and documentation for Numpy, Scipy and Matplotlib.
A great way to get to know any programming language is to solve the first few problems on project euler.
Matlab, Mex, Homebrew and OS X 10.8 Mountain Lion
Now that I am a student again, I have to use Matlab again. Among the many joys of Matlab is the compilation of mex files.
Because it does not work. So angry.
Basically, mex does not work because it assumes that you have OS X 10.6 installed. In OS X 10.6 you had gcc-4.2 and your system SDK was stored in \/Developer\/SDKs\/MacOSX10.6.sdk. However, as of 10.7 (I think), the \/Developer directory has been deprecated in favor of distributing the whole development environment within the App package of XCode. Also, gcc has been deprecated in favor of clang. While a gcc binary is still provided, gcc-4.2 is not. Of course, that is what mex relies on. Lastly, mex of course completely disregards common system paths such as, say, \/usr\/local\/bin, so compiling against some homebrew library won't work.
At least these things are rather easy to fix, since all these settings are saved in a file called mexopts.sh, which is saved to ~\.matlab\/R2012a\// by default. The relevant section on 64-bit OS X begins after maci64) and should look like this: (changes are marked by comments)
#----------------------------------------------------------------------------
# StorageVersion: 1.0
# CkeyName: GNU C
# CkeyManufacturer: GNU
# CkeyLanguage: C
# CkeyVersion:
CC='gcc' # used to be 'gcc-4.2'
# used to be '/Developer/SDKs/MacOSX10.6.sdk'
SDKROOT='/Applications/Xcode.app/Contents/Developer/Platforms/MacOSX.platform/Developer/SDKs/MacOSX10.8.sdk'
MACOSX_DEPLOYMENT_TARGET='10.8' # used to be '10.5'
ARCHS='x86_64'
CFLAGS="-fno-common -no-cpp-precomp -arch $ARCHS -isysroot $SDKROOT -mmacosx-version-min=$MACOSX_DEPLOYMENT_TARGET"
CFLAGS="$CFLAGS -fexceptions"
CFLAGS="$CFLAGS -I/usr/local/include" # Homebrew include path
CLIBS="$MLIBS"
COPTIMFLAGS='-O2 -DNDEBUG'
CDEBUGFLAGS='-g'
#
CLIBS="$CLIBS -lstdc++"
# C++keyName: GNU C++
# C++keyManufacturer: GNU
# C++keyLanguage: C++
# C++keyVersion:
CXX=g++ # used to be 'g++-4.2'
CXXFLAGS="-fno-common -no-cpp-precomp -fexceptions -arch $ARCHS -isysroot $SDKROOT -mmacosx-version-min=$MACOSX_DEPLOYMENT_TARGET"
CXXLIBS="$MLIBS -lstdc++"
CXXOPTIMFLAGS='-O2 -DNDEBUG'
CXXDEBUGFLAGS='-g'
#
# FortrankeyName: GNU Fortran
# FortrankeyManufacturer: GNU
# FortrankeyLanguage: Fortran
# FortrankeyVersion:
FC='gfortran'
FFLAGS='-fexceptions -m64 -fbackslash'
FC_LIBDIR=`$FC -print-file-name=libgfortran.dylib 2>&1 | sed -n '1s/\/*libgfortran\.dylib//p'`
FC_LIBDIR2=`$FC -print-file-name=libgfortranbegin.a 2>&1 | sed -n '1s/\/*libgfortranbegin\.a//p'`
FLIBS="$MLIBS -L$FC_LIBDIR -lgfortran -L$FC_LIBDIR2 -lgfortranbegin"
FOPTIMFLAGS='-O'
FDEBUGFLAGS='-g'
#
LD="$CC"
LDEXTENSION='.mexmaci64'
LDFLAGS="-Wl,-twolevel_namespace -undefined error -arch $ARCHS -Wl,-syslibroot,$SDKROOT -mmacosx-version-min=$MACOSX_DEPLOYMENT_TARGET"
LDFLAGS="$LDFLAGS -bundle -Wl,-exported_symbols_list,$TMW_ROOT/extern/lib/$Arch/$MAPFILE"
LDFLAGS="$LDFLAGS -L/usr/local/lib" # Homebrew library path
LDOPTIMFLAGS='-O'
LDDEBUGFLAGS='-g'
#
POSTLINK_CMDS=':'
#----------------------------------------------------------------------------
To summarize:
- changed
gcc-4.2togcc - changed
/Developer/SDKs/MacOSX10.6.sdkto/Applications/Xcode.app/Contents/Developer/Platforms/MacOSX.platform/Developer/SDKs/MacOSX10.8.sdk - changed
10.5to10.8 - added
CFLAGS="$CFLAGS -I/usr/local/include" - changed
g++-4.2tog++ - added
LDFLAGS="$LDFLAGS -L/usr/local/lib"
With those settings, the mex compiler should work and it should pick up any libraries installed by homebrew.
Kompilieren auf Windows

Seit einigen Wochen arbeite ich an einem kleinen Projekt: Eine Matlab-Funktion, die, ähnlich wie die standard-Funktion wavread(), Audiodateien einlesen kann. Aber nicht irgendwelche Audiofiles, sondern ALLE MÖGLICHEN Audiofiles. Wie geht das? Jeder kennt VLC, den Video-Player, der so ziemlich jedes Video öffnen kann, das man ihm vorsetzt, selbst wenn man überhaupt keine Codecs installiert hat. VLC basiert auf FFmpeg, einem Open-Source Programm, welches Funktionen bereit stellt, um eben alle möglichen Mediendaten zu öffnen.
Und da FFmpeg freie Software ist, kann man sie auch für andere Dinge verwenden, etwa, um mit Matlab Audiodateien zu öffnen. Fehlt noch eine Verbindung zwischen Matlab und den FFmpeg-C-Bibliotheken, und die gibt es in Form von Mex, der C-Schnittstelle von Matlab. Feine Sache, zwar hat es eine Weile gedauert, bis ich mich in libavformat und libavcodec eingearbeitet hatte (die beiden wichtigsten FFmpeg-Bibliotheken), aber im Endeffekt lief das alles sehr schmerzfrei – und das, obwohl ich bisher Mex-Kompilieren mit Matlab immer als eine grausige Beschäftigung in Erinnerung hatte, gespickt von kryptischen Kompiler-Fehlern und hässlichen Notlösungen.
Bumms, Zack, kaum hatte ich mich versehen, hatte ich ein lauffähiges, tadellos funktionierendes Mex-File auf meinem Mac liegen. Damit hatte ich nicht gerechnet. Also sofort die momentane Euphorie ausnutzen und weiter zu Schritt 2, das Ganze nochmal auf Windows. Meine Probleme, Windows so einzurichten, dass ich endlich Kompilieren kann, hatte ich ja schon berichtet. Ich hatte also Visual Studio 2005 installiert, um Matlab zufrieden zu stellen und einen anständigen Kompiler auf dem System zu haben. Aber war ja klar, MSVC macht wieder sein eigenes Ding und nichts ist mit Standardkonformität und Trallalla: Keine C99-Unterstützung, also keine Variablendeklarationen mitten im Code und keine stdint.h oder inttype.h. Ein Glück, es gibt wieder ein wenig mehr Free Software, die wenigstens letztere Lücke schließt. Dennoch; Ich bekomme mein mex-File nicht zum Laufen. Es ist wie verflucht, kaum setze ich mich an eine Windows-Maschine zum Programmieren, fällt meine Produktivität auf das Niveau eines Backsteins.
Enter gnumex, noch ein weiteres Stück FOSS, das es ermöglicht, GCC als Mex-Kompiler zu verwenden, AUF WINDOWS. Um die Dinge zu vereinfachen, verwendete ich die MinGW-Variante und kaum war diese Hürde genommen… lief alles. Einfach so. Wahrscheinlich bin ich ein Dickschädel und habe einfach nicht die Geistesschärfe, mit Windows-Kompilern zu arbeiten, aber mir scheint, alles was ich diesbezüglich anfasse und das nicht GCC heißt ist zum Scheitern verurteilt. Ein Glück, dass es die vielen klugen Jungen und Mädchen gibt, die so wunderbare freie Software schreiben, die mir das Leben so viel einfacher macht!
Eine Fortsetzung kommt noch…
Vom Sinn und Unsinn, ein Cross-Platform-Compiling-Matlab-System aufzubauen
Mal wieder: Ich schreibe ein Stück Software für meinen Nebenjob bei meinem Signalverarbeitung-Prof. Diesmal geht es darum, beliebige Audio-Files in Matlab einlesen zu können. Perfekt geeignet ist dafür die selbe Bibliothek, die auch von VLC verwendet wird, libavcodec/libavformat. Das ist eine normale C-Bibliothek, es braucht also nur noch ein kleines mex-File, um ihre Funktionalität für Matlab zur Verfügung zu stellen. Klappt auch wunderbar. Auf dem Mac.
Schritt zwei ist dann, das Ganze auf Windows und Linux zum Laufen zu bringen. Eigentlich kein Problem, denn ich habe keine wilden Dinge getan und die Libraries selbst sind wunderbar Cross-Platform, es gibt sie sogar schon vorkompiliert für praktisch jedes denkbare Betriebssystem.
Also, was brauche ich? Zwei Dinge: Matlab und einen C-Compiler (der mitgelieferte LLC-Compiler macht mein Hirn bluten). Matlab zu installieren ist meiner Erfahrung nach schmerzhaft. Bigtime. Nicht, weil Matlab schwer zu installieren wäre, sondern, weil Mathworks nur zwei Installationen pro Schachtel erlaubt, was für meine drei Betriebssysteme zu gewissen Problemen führt. Außerdem müsste ich meine eine Lizenz erst für die Windows-installation umschreiben lassen, und… ach, Schmerzen. Offenbar habe ich die Jungs dort aber schon derart häufig mit Lizenzanfragen genervt, dass sie mich einfach als hoffnungslos aufgegeben haben, denn dieses Mal musste ich keine neue Lizenz erstellen lassen, sondern einfach installieren, Passwort eingeben, und los. Mein Account meldet jetzt, dass ich fünf gleichzeitige Installationen hätte (von zwei erlaubten). Mir solls Recht sein.
Außerdem: ein aktuelles Linux muss her. VMWare sei Dank, lauert im Linux-Installieren nicht mehr der Schrecken, im Zweifelsfall den kompletten Festplatteninhalt zu verlieren, sondern nur noch, an akuter Progressbar-itis zu ersticken. War ja klar, dass Autoupdate sich diesen Nachmittag aussucht, um meine Ubuntu-VM hoffnungslos zu zerstören. Also, neues Ubuntu heruntergeladen, neu installiert, neu Updates aufgespielt, zwei Stunden Lifetime verloren. Immerhin: es hat fehlerfrei funktioniert, das ist was Neues. Matlab hinterher, VMWare Tools dazu, fertig ist die Development-Kiste. Jetzt fehlt nur noch eine Verbindung zu meinem Development-Verzeichnis, damit ich auf meine Dateien zugreifen kann. Fehlanzeige. Dukommsthiernichrein. Na Toll.
Also weiter zu Windows. Frühere Versuche ergaben bereits, dass ich Matlab nicht dazu bewegen kann, (a) GCC als Compiler zu nehmen oder (b) das bereits installierte Microsoft Visual Studio C++ .Net Professional Directors Cut Special Edition 2008 Ultimate zu verwenden. Nähere Nachforschungen zeigen: Zu neu, Kennternich. Geht nur bis MSVC Jahrgang 2005. Also: Neues MSVC deinstallieren, Altes installieren. ich freue mich immer darauf, MSVC zu deinstallieren, denn es besteht lediglich aus kompakten 12 Programmen, die sich zwar alle auf einem Haufen Installieren- jedoch nicht De-Installieren lassen. Immerhin ist es dank MSDNAA-Membership nicht schwer, an die alten Versionen heranzukommen. Und klar, die Systemsteuerung lässt einen auch immer nur ein Programm auf einmal deinstallieren. Multitasking ist nicht. Dank Syncplicity kann Windows die Zwischenzeit immerhin dazu verwenden, all meine Development-Files auf den Rechner zu laden. Yay! Das Schöne an Fortschrittsbalken ist ja, sie zeigen Fortschritt. Damit haben sie einen klaren Vorteil gegenüber etwa Dachbalken oder den Bittewartenpunktpunktpunkt-Balken, die die Microsoft SQL Server 2008-Deinstallation stolz herumzeigt. Die fühlt sich wohl sehr wichtig, denn sie rödelt eine starke halbe Stunde im Bitte-Warten-Modus herum. So mag ich Deinstallationen.
To be continued…

bastibe.de by Bastian Bechtold is licensed under a Creative Commons Attribution-ShareAlike 3.0 Unported License.